Manuscript accepted on : 19-06-2025
Published online on: 23-06-2025
Plagiarism Check: Yes
Reviewed by: Dr. Vinutha K B
Second Review by: Dr. Nagham Aljamali
Final Approval by: Dr. Wagih Ghannam
Gene Scanning for sY242 Microdeletion in Males with Infertility
Mansi Dadhania , Shivani Patel
, Shivani Patel and Jenabhai Chauhan*
and Jenabhai Chauhan*
Department of Genetics, Ashok and Rita Patel Institute of Integrated Study and Research in Biotechnology and Allied Science (ARIBAS), The Charutar Vidya Mandal University, Vallabh Vidyanagar, Anand, Gujarat.
Corresponding Author Email: jenabhaichauhan@aribas.edu.in
DOI : http://dx.doi.org/10.13005/bbra/3406
ABSTRACT: Infertility is a significant disorder of the male or female reproductive system, defined by the failure to conceive after twelve months or more of unprotected sexual intercourse. Around 9% of males and 10% of females aged 22-44 reported trouble in reproducing. Y-chromosome microdeletions within the azoospermia factor (AZF) regions represent a major genetic cause of male infertility. In the current study, sperm morphological examination was performed using Papanicolaou staining to identify spermatozoa abnormalities. Gene scanning for sY242 (AZFc sub-region) microdeletion was performed by STS-PCR. Total of 100 semen samples (67 normozoospermia, 30 oligozoospermia and 3 azoospermia including blood) were examined for sperm morphological abnormalities and Y chromosomal microdeletion. Eighty-seven samples showed variable amount of sperm defects whereas 13 samples showed high amount of head, tail and mid piece defects. Gene scanning at sY242 using STS-PCR technique showed microdeletion in the 13 samples which includes 5 normozoospermic (7.46%), 7 oligozoospermic (23.33%) and 1 Azoospermic (33.3%) males. Our findings suggest that there is a involvement of sperm morphological defects during sY242 microdeletion in the studied infertile males. However, the study can be extended for more number of samples to rule out actual frequency of microdeletion at sY242 gene and its association with sperm morphological defects. This study will be of great help to infertility clinics for genetic counseling and assisted reproduction.
KEYWORDS: Gene Scanning; Male infertility; Microdeletions; STS-PCR; Y chromosome
Download this article as:| Copy the following to cite this article: Dadhania M, Patel S, Chauhan J. Gene Scanning for sY242 Microdeletion in Males with Infertility. Biotech Res Asia 2025;22(2). |
| Copy the following to cite this URL: Dadhania M, Patel S, Chauhan J. Gene Scanning for sY242 Microdeletion in Males with Infertility. Biotech Res Asia 2025;22(2). Available from: https://bit.ly/4ecsq4t |
Introduction
Infertility is described as the reproductive incapacity to conceive pregnancy after 365 days or more of unprotected sexual contact. This condition affects 10%-15% of couples globally, with male-related variables accounting for half of the cases.1
Hormonal disorders, erectile malfunction, infections, anti-sperm antibodies, contact with chemical agents and radiations, testicular cancer, varicocele, hereditary variables, and many more have all been linked to male infertility. As a result, male infertility is a complex condition that encompasses a wide range of problems. Yet, the cause of infertility is unclear in around 30%-50% of male cases.2
Forty to Fifty (40-50%) of fertile males have sperm production problems such as sperm quantity decrease, motility, or aberrant morphology. Yet, the root cause of infertility is still unknown. Male infertility is thought to be caused by autosomal and sex chromosomal abnormalities in 10- 15% of case.3
Y Chromosome and Male Infertility
Deletions in the Azoospermia Factor (AZF) regions of the human Y chromosome are recognised as a prevalent cause of severe testicular dysfunction and spermatogenic abnormalities resulting in male infertility. It is commonly accepted that at least three nonoverlapping sections of the human Y chromosome—AZFa, AZFb, and AZFc—are essential for normal spermatogenesis.4 However, there is still limited data available for Indian infertile men.5
AZFc region is located at distal end of interval 6 and spans around 3.5 Mb. Deletion of these regions observed some azoospermic and sever oligozoospermic patients. AZFc contain eight copies gene; namely: DAZ, BPY2. CDY1, CSPG4, LYP1, TTTY3, TTTY4 and TTTY17 which are expressed only in testis.6 AZFc deletion is caused by recombination involving b2/b4 sequences and deleted all 4 copies of DAZ gene and 3 copies of BPY2 genes.7 Genes associated with AZFc region are sY254, sY255, sY145, sY152, sY153, sY157, sY239, sY242, sY802, sY856, sY1191, sY2713, sY2900, sY2928.8
As infertility is a global problem, present study was taken to scan sY242 microdeletion in males with fertility problem.
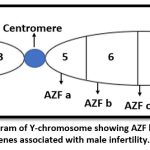 |
Figure 1: Illustrative Diagram of Y-chromosome showing AZF loci and their candidate genes associated with male infertility. |
Materials and methods
Sample selection
Total of 100 semen samples were collected from different District of Gujarat. Among the 100 samples, 67 were normozoospermia, 30 were oligozoospermia and 3 were azoospermia. The consent form containing the details about research purpose and methods which was given to the male participants for their information and consent. Only Males who signs the consent form, their samples were collected. Blood samples from Azoospermic males also collected.
Study Procedure
Genomic DNA from semen & blood samples were isolated from the samples using phenol/chloroform method. Analysis of microdeletion at sY242 of human Y-chromosome was carried out by using polymerase chain reaction (PCR). Using specific primer sets, PCR amplification was performed. For both the STS markers (SRY, sY242) independent PCR reactions were designed to analyze the microdeletions in AZFc sub-region of Yq chromosome. Polymerase chain reaction (PCR) was carried out in a 25 µL reaction volume (Table 1), containing 5 µL of genomic DNA. Amplification was performed using a HIMEDIA thermal cycler with an initial denaturation step at 94 °C for 4 minutes, followed by 34 cycles of denaturation at 94 °C for 30 seconds, annealing at marker-specific temperatures (Table 2), and extension at 72 °C for 40 seconds. A final extension was performed at 72 °C for 3 minutes, followed by a hold at 4 °C. PCR amplicons were resolved on a 2% agarose gel, stained with ethidium bromide (EtBr), visualized under a UV transilluminator. In case of presence of microdeletions in STS, PCR was repeated thrice to confirm the deletions.
Table 1: Amount (µL) of PCR components per reaction
| STSmarkers | Master Mix (2X) | Nuclease freewater | Forward primer(10 pM) | Reverse primer(10 pM) | DMSO | DNA |
| sY14 | 12.5 | 5.4 | 1 | 1 | 0.1 | 5 |
| sY242 | 12.5 | 6.4 | 0.5 | 0.5 | 0.1 | 5 |
Table 2: Primer details.9
| Region | STSMarker | Primer Sequence (5’-3’) | ExpectedAmplicon size (bp) | AnnealingTemperature (0C) |
| SRY | sY14 | F-GAATATTCCCGCTCTCCGGA | 472 | 56 |
| R-GCTGGTGCTCCATTCTTGAG | ||||
| AZFc | sY242 | F- ACA CAG TAG CAG CGG GAG TTA CR- TCT GCC ACT AAA CTG TAA GCT CC | 233 | 60 |
Results
A total of one hundred semen samples were collected from various laboratories of Gujarat. Details were obtained from the location of collection regarding the regular examination of semen samples, such as volume, colour, viscosity, motility, concentration, etc. The males were mostly between the ages of 25 and 35 years, with an age range of 21 to 40 years. The seminograph results, particularly the sperm counts, were used to classify the samples into three groups: Normozoospermia, Oligozoospermia, and Azoospermia. In all these samples, 67(67%) samples were Normozoospermia, 30(30%) samples were Oligozoospermia and 03(3%) samples were of Azoospermia. The distinct ID number was written on the samples’ labels.
Analysis of sperm morphology is completed using a Papanicolaou staining kit, several sperm morphological abnormalities (Head defects, midpiece defects and tail defects) are detected under a light microscope. Among one hundred samples, thirteen samples had a high frequency of head, tail, and midpiece defects in sperm, whereas 87 samples had various types of sperm defects which are acceptable as per the guideline.10
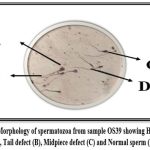 |
Figure 2: Morphology of spermatozoa from sample OS39 showing Head defect (A), Tail defect (B), Midpiece defect (C) and Normal sperm (D). |
Semen and blood samples were treated using regular phenol-chloroform techniques to isolate DNA. Qualitative and quantitative analysis of isolated genomic DNA carried out to check the the quality of extracted DNA, 1% agarose gel electrophoresis was done. Sharp, uninterrupted bands of male DNA were seen under the UV transilluminator. The electrophoresis pattern of extracted genomic DNA is presented in Figure 3.
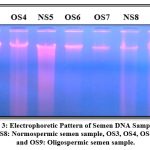 |
Figure 3: Electrophoretic Pattern of Semen DNA Sample. NS5 and NS8: Normospermic semen sample, OS3, OS4, OS6, OS7 and OS9: Oligospermic semen sample. |
An STS for SRY (sY14), which is located on Yq, is used to verify the existence of the Y chromosome and differentiate between a negative result and a technical malfunction. The SRY gene indicates that DNA is present on the Y chromosome, which is of 472 bp as shown in Figure 4, and the result bands were falling inside the ladder designated zone near to 500 bp when the PCR products were observed under 2% agarose gel.
 |
Figure 4: PCR products of representative SRY gene (472bp). NS17, NS47 and NS67: Normospermic semen sample, OS20, OS39 and OS57: Oligospermic semen sample and L: DNA Ladder (100 bp). |
In 100 infertile males, Gene scanning at sY242 using STS-PCR technique showed microdeletion in the 13 samples which includes 5 normozoospermic (7.46%), 7 oligozoospermic (23.33%) and 1 Azoospermic (33.3%) males (Table:3). While observing the PCR products under 2% agarose gel, the product bands fell inside a ladder-marked zone between 200-300 bp bands indicating the normal sY242 gene with 233bp shown in Figure 5.
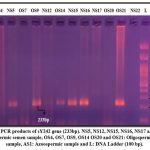 |
Figure 5: PCR products of sY242 gene (233bp). NS5, NS12, NS15, NS16, NS17 and NS22: Normospermic semen sample, OS4, OS7, OS9, OS14 OS20 and OS21: Oligospermic semen sample, AS1: Azoospermic sample and L: DNA Ladder (100 bp). |
Table 3: Gene scanning for sY242 microdeletion at AZFb region.
| Samples Types | No. of samples | % of sample with morphological defects and microdeletion | % of sample with low sperm count |
| Azoospermic | 03 | 33.3 (n=1) | – |
| Oligozoospermic | 30 | 23.33 (n=07) | 13.33 |
| Normozospermic | 67 | 7.46 (n=05) | 00 |
Discussion
According to previous research report, three out of four patients had AZF deletions that included the AZFc region, resulting in a significant portion of AZF deletions. Our results match with previously released information. 3.85% (3 out of 78) of azoospermic males had AZF deletions when the patients were grouped according to their spermiogram.11 While deletions of the AZFc area can result in varying degrees of spermatogenic failure, from oligozoospermia to the loss of germ cells in the testis, deletions of the AZFa and AZFb regions are often linked to azoospermia and failure of testicular sperm retrieval.12
It has also been discovered that the AZFc region subdeletions gr/gr, b2/b3, b1/b3, and b2/b4 are linked to male infertility.13,14 They studied Indian, Poland, Vietnam, US and Sri Lankan populations, with the prevalence of b2/b3 deletion range from 0.5 percent in India to 2.2 percent in Poland, and the incidence of gr/gr deletion ranging from 2 percent in the US to 15 percent in Vietnam. On the other hand, the frequency of b1/b3 and b2/b4 deletions did not significantly changes.15
Available data as compared to deletions in the AZFa and AZFb areas, deletions in the AZFc region are more prevalent.16 According to past studies, 4/50 (8.0%) of the male infertile patients had deletions in the AZFc region.17
Our findings concur with Lin et al18, who found 8.5% of AZFc deletions in 94 male infertile subjects analysed in Taiwan. Italian researchers found that 6.5% of male infertiles had AZFc deletions.19 However, modest frequencies 15% and 19.9% of infertile men from the Egyptian and Russian, respectively, with AZFc deletions were reported.20,21
According to earlier research, 24 people or 82.8% of the total number of azoospermic men had deletions in the AZFc region.22 This is consistent with previous research that found that, in comparison to the AZFa and AZFb areas, the AZFc region had a higher prevalence of deletion.23,24 Researcher showed that there is a higher risk of subfertility and infertility associated with a reduction in the copy number of DAZ genes (AZFc). Seven people (24.1%) had AZFc alone (together with DYZ) out of the total 29 deletions. It was intriguing to see that Y chromosomes in total 8 (27.6%) had the heterochromatic region (DYZ) deleted; this Y chromosome was identified as being short.25
Ambulkar and Pande (2018) studied microdeletions on Y chromosome through multiplex PCR utilizing 11 STS markers excluding sY242 in Indian infertile males showed that the AZFc region has the most deletions sites (10%) followed by AZFb (6.36%) and AZFa (1.81%).26
Partial deletions of the AZFc region were studied for the first time in a small samples of men from Sri Lanka. The gr/gr pattern was the most often seen partial AZFc deletion pattern. It was found in four (4.2%) patients and four (4.6%) control males, suggesting that there was no difference in the frequency of gr/gr deletions between the patient and control groups26. We also found microdeletion in the 7.4% (5/67) control samples which are 1.6 times more than that was reported by Fenando et al27 in 2006. Their studies from Sri Lanka support earlier discoveries made in other populations, which showed that spermatogenetic failure cannot be caused just by incomplete AZFc region deletions.27
Conclusion
In the current study, deviations in spermatozoa were identified by sperm morphological evaluation using Papanicolaou staining. STS-PCR was used to scan the genes for the microdeletion of sY242 (AZFc sub-region). A total of 100 semen samples (67 normozoospermia, 30 oligozoospermia, and 3 azoospermia including blood) were analyzed to check for Y chromosomal microdeletion and abnormalities in the sperm morphology. Thirteen samples had a high frequency of head, tail, and midpiece defects, whereas eighty-seven samples had varying levels of sperm defects, which are acceptable as per WHO manual 2021. Using the STS-PCR approach, Gene scanning at sY242 using STS-PCR technique showed microdeletion in the 13 samples which includes 5 normozoospermic (7.46%), 7 oligozoospermic (23.33%) and 1 Azoospermic (33.3%) males. According to our research, the infertile males under study may have had sperm morphological abnormalities during sY242 microdeletion. To rule out the true incidence of microdeletion at the sY242 gene and its correlation with sperm morphological abnormalities, the investigation might be expanded to including a larger number of samples. This study will be of great help to infertility clinics for genetic counseling and assisted reproduction.
Acknowledgement
The authors wish to thank the Head (I/C) of Ashok and Rita Patel Institute of Integrated Study & Research in Biotechnology and Allied Sciences (ARIBAS), Vallabh Vidyanagar and CVM University for providing a platform for this research work. The authors also thankful to the laboratories and hospitals staff for their direct or indirect help and support. This work was supported in part by SHODH- scheme of developing High quality research, Education department, Gujarat State.
Funding Sources
The author(s) received no financial support for the research, authorship, and/or publication of this article.
Conflict of Interest
The authors do not have any conflict of interest.
Data Availability Statement
This statement does not apply to this article.
Ethics Statement
The present study was ethically approved from the institutional ethics committee of the Govindbhai Jorabhai Patel Ayurveda Hospital and Maternity Home in Gujarat, India, (No. CVMU/GJPIASR/IEC/06/2022-23/03).
Informed Consent Statement
The consent form containing the details about research purpose and methods which was given to the male participants for their information and consent. Only Males who signs the consent form, their samples were collected. The privacy rights of human subjects must always be observed.
Clinical Trial Registration
This research does not involve any clinical trials.
Permission to reproduce material from other sources
Not Applicable
Authors Contributions
Mansi Dadhania: Conceptualization, Materials and Methodology, Analysis, Writing- Original Draft.
Shivani Patel: Analysis, Data collection.
Jenabhai Chauhan: Visualization, Supervision, Reviewing and editing Draft.
References
- World Health Organization, Infertility. WHO. Published September 14, 2023. Accessed April 28, 2025. https://www.who.int/news-room/fact-sheets/detail/infertility
CrossRef - Zhang, Z., Xi, Q., Wang, R., et al. Obstetric and perinatal outcomes of intracytoplasmic sperm injection for infertile men with Y chromosome Medicine. 2019;98(41): e17407. DOI:10.1097/ MD.0000000000017407
CrossRef - Anannya G., Rita M., Purnali N., et al. Analysis for the detection of frequency of Y chromosome microdeletions in the AZFa, AZFb and AZFc region in different infertile groups of North East Indian Population. International Journal of Scientific Research. 2017;6(4):2277 –1
- Reijo, R., Alagappan, K., Page, C., & Patrizio, P. Severe oligozoospermia resulting from deletions of azoospermia factor gene on Y chromosome. The Lancet. 1996;347(9011):1290-1293. DOI: 1016/S0140-6736(96)90938-1
CrossRef - Rani, S., Rajender, S., Pavani, K., et al. High frequencies of Non Allelic Homologous Recombination (NAHR) events at the AZF loci and male infertility risk in Indian Scientific Reports. 2019;9(1):6276. DOI https://doi.org/10.1038/s41598-019-42690-0
CrossRef - Eloualid, A., Abidi, O., Charif, M., et al. Association of the MTHFR A1298C variant with unexplained severe male infertility. PloS one. 2012;7(3), e34111. https://doi.org/10.1371/journal.pone.0034111
CrossRef - Yu, XW., Wei, ZT., Jiang, YT., & Zhang, L. Y chromosome azoospermia factor region microdeletions and transmission characteristics in azoospermic and severe oligozoospermic patients. International journal of clinical and experimental medicine. 2015;8(9):14634-14646. PMID: 26628946 PMCID: PMC4658835
- Jia, C., Li, L., Chen, S., et al. Cytogenetic and molecular characterization of an oligoasthenozoospermia male carrier of an unbalanced Y; 22 translocation: a case Medicine. 2019;98(15):e15209. DOI: 10.1097/MD.0000000000015209
CrossRef - Krausz, C., Hoefsloot, L., Simoni, M., & Tuttelmann, F. EAA/EMQN best practice guidelines for molecular diagnosis of Y‐chromosomal microdeletions: state‐of‐the‐art 2013. Andrology. 2014;2(1):5-19. https://doi.org/10.1111/j.2047-2927.2013.00173.x
CrossRef - World Health Organization. WHO Laboratory Manual for the Examination and Processing of Human Semen. 6th ed. World Health Organization; 2021. Accessed April 28, 2025. https://www.who.int/ publications-detail-redirect/9789240030787
- Suganya, J., Kujur, S., Selvaraj, K., Suruli, M., Haripriya, G., & Samuel, C. Y chromosome microdeletions and partial AZFc deletions in infertile men from South India. British Journal of Medicine and Medical Research. 2016;13(12):1-10. DOI: 10.9734/BJMMR/2016/24208
CrossRef - Foresta, C., Moro, E., & Ferlin, A. Y chromosome microdeletions and alterations of spermatogenesis. Endocrine reviews. 2001;22(2):226-239. https://doi.org/10.1210/edrv.22.2.0425
CrossRef - Repping, S., Skaletsky, H., Brown, L., et al. Polymorphism for a 1.6-Mb deletion of the human Y chromosome persists through balance between recurrent mutation and haploid selection. Nature genetics. 2003;35(3):247-251. DOIhttps://doi.org/10.1038/ng1250
CrossRef - Fernandes, S., Paracchini, S., Meyer, L., Floridia, G., Tyler-Smith, C., & Vogt, P. A large AZFc deletion removes DAZ3/DAZ4 and nearby genes from men in Y haplogroup N. The American Journal of Human Genetics. 2004;74(1):180-187. DOI: 1086/381132
CrossRef - Rozen, G., Marszalek, D., Irenze, K., et al. AZFc deletions and spermatogenic failure: a population-based survey of 20,000 Y chromosomes. The American Journal of Human Genetics. 2012;91(5):890-896. DOI: 1016/j.ajhg.2012.09.003
CrossRef - Sawai, H., Komori, S., & Koyama, K. Molecular analysis of the Y chromosome AZFc region in Japanese infertile males with spermatogenic defects. Journal of reproductive immunology. 2002;53(1-2):37-44. https://doi.org/10.1016/S0165-0378(01)00090-0
CrossRef - Swarna, M., Babu, R., & Reddy, P. AZFc deletions in idiopathic infertile males from south India. International Journal of Human Genetics. 2003;3(1):1-4. https://doi.org/10.31901/ 24566330.2003/03.01.01
CrossRef - Lin, M., Chen, W., Sun, S., et al. Y-chromosome microdeletion and its effect on reproductive decisions in Taiwanese patients presenting with nonobstructive azoospermia. Urology. 2000;56(6):1041-1046. https://doi.org/10.1016/S0090-4295(00)00846-3
CrossRef - Ferlin, A., Moro, E., Garolla, A., & Foresta, C. Human male infertility and Y chromosome deletions: role of the AZF-candidate genes DAZ, RBM and DFFRY. Human reproduction. 1999;14(7):1710-1716. https://doi.org/10.1093/humrep/14.7.1710
CrossRef - Eid, M., Eid, M., Abdelrahman, A., et al. Detection of AZFc gene deletion in a cohort of Egyptian patients with idiopathic male infertility. Journal of Genetic Engineering and Biotechnology. 2023;21(1):111. https://doi.org/10.1186/s43141-023-00584-9
CrossRef - Osadchuk, V., Vasiliev, V., Ivanov, K., et al. Prevalence of AZFс Y chromosome microdeletions and association with spermatogenesis in Russian men from the general population. Vavilov Journal of Genetics and Breeding. 2024;28(7):780. doi:10.18699/vjgb-24-86
CrossRef - Thangaraj, K., Gupta, J., Pavani, K., et al. Y chromosome deletions in azoospermic men in India. Journal of andrology. 2003;24(4):588-597. https://doi.org/10.1002/j.1939-4640.2003.tb02710.x
CrossRef - Martinez, C., Bernabés, J., Gomez, E., et al. Screening for AZF deletion in a large series of severely impaired spermatogenesis patients. Journal of andrology. 2000;21(5):651-655. https://doi.org/10.1002/j.1939-4640.2000.tb02132.x
CrossRef - Peterlin, B., Kunej, T., Sinkovec, J., Gligorievska, N., & Zorn, B. Screening for Y chromosome microdeletions in 226 Slovenian subfertile men. Human reproduction. 2002;17(1):17-24. https://doi.org/10.1093/humrep/17.1.17
CrossRef - de Vries, W., Hoffer, J., Repping, S., Hoovers, M., Leschot, J., & van der Veen, F. Reduced copy number of DAZ genes in subfertile and infertile men. Fertility and sterility. 2002;77(1):68-75. https://doi.org/10.1016/S0015-0282(01)02935-1
CrossRef - Ambulkar, P. S., & Pande, S. S, Study of Y-chromosome microdeletions in azoospermic infertile males using multiplex PCR analysis. Biosciences Biotechnology Research Asia, 2018;15(2):351.
CrossRef - Fernando, L., Gromoll, J., Weerasooriya, R., Nieschlag, E., & Simoni, M. Y‐chromosomal microdeletions and partial deletions of the Azoospermia Factor c (AZFc) region in normozoospermic, severe oligozoospermic and azoospermic men in Sri Lanka. Asian journal of andrology. 2006;8(1):39-44. https://doi.org/10.1111/j.1745-7262.2006.00100.x
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





