Manuscript accepted on : 12-07-2025
Published online on: 22-07-2025
Plagiarism Check: Yes
Reviewed by: Dr. Bhavesh Hirabhai Patel
Second Review by: Dr. Supriya Shivanand Mhamane
Final Approval by: Dr. Eugene A. Silow
Kalaivani Devaraj1,2* and Dheepa Ganapathy 1
and Dheepa Ganapathy 1
1Department of Computer Science, PKR Arts College for Women, Tamilnadu, India
2Department of Computer Science, PPG College of Arts and Science, Tamilnadu, India
Corresponding author Email: kalairathin@gmail.com
ABSTRACT: To propose a hybrid quantum-based deep learning model to detect malignant lung nodules and accurately classify disease severity. Bio-inspired techniques are integrated to optimize the learning rate for robust generalization. A Hybrid Quantum-Enhanced Deep Neural Network (QE-DNN) combined with a Quantum Convolutional Neural Network (Q-CNN) is used to extract the multi-scale spatial patterns from high-resolution CT-DICOM images. To perform deep segmentation, Quantum Mask-RCNN is used to isolate the ROI from the images effectively. A bio-inspired Adaptive Firefly-Differential Evolution (AFDE) optimizer is employed to fine-tune the learning architecture. Quantum histogram equalization and wavelet fusion are incorporated as data pre-processing methods to retain critical edge and intrinsic features. CT-DICOM dataset is used for evaluation which consists of 25,1135 images with a resolution of 512x512 pixels. The performance is assessed in MATLAB, TensorFlow Quantum, and IBM Qiskit tools by comparing the proposed work with existing models such as SVM-WSS, GCPSO-PNN, 3D-DLCNN, and ECNNDE-BCE. The proposed QEDNN-AFDE quantum bio-inspired nodule detection strategy enhanced the generalization capability by exploring wider with proven results of 96.4% accuracy, 95.2% sensitivity, 95.8% specificity, 95.2% F1 score, 94.6% dice coefficient, 0.02 Log Loss and AUC-ROC with 0.95 TPR and 0.05 FNR. The proposed QEDNN-AFDE model strengthens the interpretability of deep learning models in medical imaging and sets a new benchmark in quantum-assisted diagnostics in precision oncology. The model shows promising performance in both classification accuracy and severity prediction and outperforms all existing models.
KEYWORDS: Classification; Data Processing, Image Segmentation; Lung Nodules Detection; Mask R-CNN; Quantum Convolutional Neural Network
| Copy the following to cite this article: Devaraj K, Ganapathy D. Quantum Enhanced Deep Learning Bio Inspired Model for Lung Tumors Detection and Severity Analysis in Clinical CT-DICOM Images. Biotech Res Asia 2025;22(3). |
| Copy the following to cite this URL: Devaraj K, Ganapathy D. Quantum Enhanced Deep Learning Bio Inspired Model for Lung Tumors Detection and Severity Analysis in Clinical CT-DICOM Images. Biotech Res Asia 2025;22(3). |
Introduction
Lung cancer is one of the most fatal malignancies, and it is a leading cause of cancer-related deaths globally, often diagnosed at advanced stages. The severity of lung cancer ranges from localized to aggressive metastatic tumors. Early detection with accurate severity grading is crucial for better treatment strategies, prognosis, and survival outcomes in non-small and small-cell lung cancers. As we are in AI and medical imaging technological advancements, deep learning methods have shown high potential in early-stage automated tumor detection and classification by extracting multiple features. However, the prevailing techniques can detect tumors but often struggle with feature ambiguity, poor generalization, and suboptimal segmentation in heterogeneous clinical datasets. To address the limitations of prevailing models, this research proposes a hybrid deep learning quantum framework using a Quantum-Enhanced Deep Neural Network with Adaptive Firefly-Differential Evolution (QEDNN-AFDE) to detect and classify lung tumors and severity analysis with high accuracy. This approach’s uniqueness lies in quantum-inspired amplitude encoding, quantum convolutional layers, and evolutionary optimization to enhance the learning rate and diagnostic precision. The proposed work is compared against SVM-WSS, GCPSO-PNN, 3D-DLCNN, and ECNNDE-BCE models. The key contributions of this study include Novel QEDNN architecture for enhanced spatial representation, AFDE for adaptive hyperparameter tuning, deployment of quantum-enhanced Mask-RCNN for deep segmentation, and quantum pre-processing methods. This research is tailored for real-world clinical integration, which is scalable and high-precision and sets a benchmark for lung cancer diagnostics in the healthcare sector.
A deep CNN model with a dual attention mechanism was proposed to enhance lung nodule detection from CT-MICRO images. The model outperforms with a high sensitivity ratio, detecting subtle nodule patterns in dense regions. However, deep CNN architecture lacks adaptability and generalization in multi-class scenarios for diverse anatomical structures. Also, the study does not include an optimization strategy, limiting diagnostic completeness.1 Hybrid quantum architecture combined with radiographs and CT images was introduced to detect lung cancer at an early stage. In this model, quantum noise processing is employed to improve the classification accuracy. The model lacks a dedicated segmentation mechanism, which limits the spatial precision in extracting ROI in images. The authors mainly focused on binary classification without addressing the severity grades, which leads to a limited scope beyond boundary diagnosis.2 ExtRANFS, an automated malignancy detection mechanism with a random feature selection method, is proposed to improve tumor detection accuracy. However, the model highly depends on handcrafted features, limiting robustness and scalability across diverse patient profiles. End-to-end segmentation is limited in this model as it lacks clinical applicability and interpretability.3 Standard CNN deep learning model differentiates the cancer stages and severity levels. The CNN layers deeply processed the feature maps and performed based on threshold test runs. This model’s limitations lie in ROI localization and hyperparameter tuning, which is crucial for multi-class tasks.4 VGG-16 and the traditional CNN model were evaluated for lung cancer detection from CT scans. The model outperforms with 85% classification accuracy, which lacks robust optimization towards deep medical image analysis and region-specific learning capabilities. The absence of pre-processing enhancements affects the model’s sensitivity, leading to noise inputs to the segmentation and masking system.5 The IWD-ARP bio-inspired optimization model was proposed to demonstrate the utility of water drops in the decision-making process. The concept inspired the adoption of the nature-inspired algorithm in lung cancer prediction to tune the deep hyperparameters, which is central to this proposed research work.6 The SVM-WSS classical approach was developed to predict lung cancer using structured datasets. A three-stage approach is followed in the research to attain multi-class objectives. 85% accuracy is achieved by the model, which is unsuitable for critical diagnostic applications. A few limitations noted in this model are the lack of segmentation, adaptability, and optimization, which is crucial for a high-performing multi-class decision support system.7
GCPSO-PNN lung cancer segmentation and classification bio-inspired model is introduced to improve the ROI selection and to work on complex datasets. The classification uses CNN layers, incorporating edge selection and a boundary marking strategy. The model’s optimization process is completely lacking, leading to inadequate feature learning and faster convergence in real-time applications.8 A 3D-DLCNN model with a DICOM dataset was proposed to target three classes and was implemented successfully using spatial and temporal feature extraction. Though the model attained 86% accuracy, it lacks memory constraints and adaptive optimization, limiting scalability and convergence. The data distribution between layers is highly impacted due to its fixed learning parameters.9 ECNNDE-BCE1 was proposed to enhance lung nodule detection and classification. The model uses the BCE loss function to improve classification and minimize false alarms. The model’s segmentation phase delivers high-precision output, leading to a better decision-making system. The model’s limitation lies only in quantum precision and quantum layer segmentation, which are crucial for high interpretability for clinical use.10
Table 1: Literature Analysis of Specific State-of-Art Models
| Name of the Authors & Model | Methods Adapted | Merits | Demerits | Limitations of Lung Cancer Detection Multi-Class Classification |
| Wei et.al 11 has done Quantum CNN Survey | Quantum Machine Learning based convolutional layers | Employment quantum methods for imaging | Lacks optimization and deep segmentation | No hyperparameter tuning for better optimization accuracy |
| Nithyanandh et.al 12 proposed EAB-IFPA |
Improved Firefly bio-inspired optimization |
Deep optimization and hyper parameter tuning process | Limited to specific number of datasets | Doesn’t address medical image diagnosis for complex datasets |
| Ahmad and Alqurashi 13 introduced DL Cancer Detection | Deep learning models with image processing | Highlights early detection | It is Conceptual and lacks CT data trials | Lacks empirical medical tests and convergence |
| Bharathi and Shalini 14 proposed Hybrid Attention-DL method | Attention + Heuristic DL method | Strong attention mechanism focuses on specific tumor zones | Lacks generalizability beyond fusion | Not suitable for real-time diagnostics |
| Jaiganesh and Nithyanandh 15 utilized Virus Swarm algorithm | Virus Swarm Bio-Inspired |
Effective optimization | Not suitable for real-world application | Doesn’t work for complex 2D and 3D datasets |
| Elhassan et.al 16 developed Dual-Model DL |
CNN + Classifier Fusion | Fusion improves classification | Binary classification only | No stage detection support |
| Choudhury et.al 17 introduced TL-Based CT Detection | Transfer Learning with CNN approach | Efficient CT-based transfer accuracy | No dynamic segmentation layer | Limited generalizability |
| Nithyanandh and Jaiganesh 18 employed ABC-based ML model | ABC Bio-Inspired algorithm | Fast search process locally and globally | No clinical trials were done | More focus on text datasets |
| Sharma et.al 19 proposed Lung-Colon DL | Deep CNN Models | High performance for multi-cancer | No severity grading | Fails on overlapping lesions |
| Arularasan et.al 20 proposed DL Image Recognition | Deep Learning Feature Recognition | Boosts SLR accuracy | No multi-class adaptability | Computational complexity and limited feature analysis |
| Devi et.al 21 introduced GANs + Bio-Inspired Model | GANs based deep learning model | Robust mechanism to reduce data loss | No clinical image validation | Lacks on severity grading and multi-class analysis |
| Abe et.al 22 utilized Robust DL CT | Enhanced CNN from CT images |
Handles varied CT image quality | No optimization layer | No dual-task output |
| Crasta et.al 23 developed Novel DL CT Arch Model | Custom CNN + Feature Maps | Custom-tailored for tumor zones | No quantum features | No use of quantum-inspired design |
| Prabhu et.al 24 proposed Bio Swarm Routing | Virus Swarm Bio-Inspired |
Adaptive to network changes | Dynamic pre-processing |
Local and Global feature analysis |
| Hroub et.al 25 used Explainable DL | Explainable CNNs | Justified predictions via explainability | Lacks segmentation | No severity score predicted |
| Selvam and Joy 26 employed AEN + Mask-RCNN | Autoencoder + Mask-RCNN | Effective plant segmentation technique adapted | Non-lung domain | Different dataset domain |
| Nithyanandh 27 YOLOv8 + CNN | YOLOv8, Deep CNN | Real-time object identification | Focused on object tasks only | Lacks severity analysis |
| Gao et.al 28 introduced DL Annotation | Robust DL + Incomplete Labels | Improves learning with missing data | Limited to partial labels | Needs label completion tools |
| Li et.al 29 utilized Dual Stream HNN | Hybrid Neural Network | Efficient dual-path inference | No explainability module | Not suitable for DICOM imaging |
| Kwon et.al 30 proposed model for Epitope Prediction | Epitope Deep Learning Classifier | Predicts feature for allergies | Image analysis is limited | Deep segmentation and binary classification |
Materials and Methods
The proposed model focused on a novel quantum-based deep learning method integrated with a bio-inspired optimization learning mechanism to detect lung tumors and highly accurately analyze the severity score using CT-DICOM images.
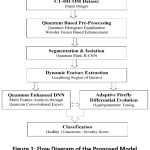 |
Figure 1: Flow Diagram of the Proposed Model |
Quantum Enhanced Deep Neural Network (QE-DNN), Quantum Mask-RCNN (QM-RCNN), and Adaptive Firefly-Differential Evolution (AFDE) methods are used for feature analysis, deep segmentation, and hyperparameter tuning and optimization. The proposed framework utilizes quantum-enhanced pre-processing, enabling the model to detect subtle tumor characteristics better. The model aims to enhance diagnostic precision and clinical interpretability using intelligent, quantum-based bio-inspired architecture. Figure 1 shows the flow diagram of the quantum hybrid framework and its methodology.
CT-DICOM Data Description and Partition Strategy
This research study utilizes the CT-DICOM dataset with 251135 images collected from the TCIA repository with tumor presence, severity scores, and bounding boxes annotations. All the images with standard pixel size 512×512 include greyscale combinations. The dataset is partitioned for training, testing, and validation purposes with a ratio of 70:20:10. Table 2 shows the dataset attributes and values with major features used for this research work to meet the objective. The data splitting is done exclusively for training, tuning, and unbiased end assessment.
Table 2: Dataset Description and Features
| Attributes | Values |
| Total Number of Datasets | 251135 |
| Number of Training Data | 175794 |
| Number of Testing Data | 50227 |
| Number of Validating Data | 25114 |
| Number of Major Features | 20 |
| Number of Major Features Extracted | |
| Image_ID , Pixel_Intensity_Distribution , Texture_Entropy , Nodule_Size , Shape_Descriptors , Lung_Lobe_Location , Bounding_Box_Coordinates , Gray_Level_Co-occurrence_Matrix (GLCM), Histogram_Equalization_Values , Wavelet_Coefficients , Segmentation_Mask_Accuracy , Edge_Sharpness_Index , Mean_Intensity, Standard_Deviation Skewness , Kurtosis , Gradient_Orientation , Quantum_Encoded_Feature_Vector
Class_Label (0: Healthy, 1: Cancer, 2: Severity) |
|
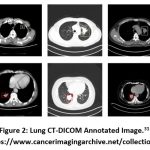 |
Figure 2: Lung CT-DICOM Annotated Image.31 |
Quantum-Enhanced Image Pre-Processing using QHE and WEF
In order to enhance the raw data quality and boost the robust feature learning data-preprocessing is required. In this research study, quantum enhanced image processing pipeline is employed to refine the CT-DICOM images prior to segmentation and classification stage. The pre-processing consists of four stages such as i) intensity normalization ii) histogram equalization iii) wavelet-based noise removal and iv) quantum-based feature transformation.
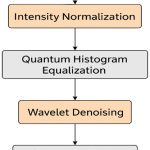 |
Figure 3: Pre-Processing of CT-DICOM Images |
For intensive normalization, the variability is measured in HU (Hounsfield Units) across every CT-DICOM, the images are normalized to standard intensity ranges. The min-max normalization technique is applied for each image pixel Ii,j. The normalization equation is expressed in Equation 1.
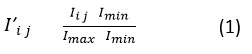
where, Imax, Imin represents minimum and maximum HU values in the DICOM input image. This method ensures greater consistency in greyscale representations across datasets which is highly essential for downstream quantum encoding. In order to improve the contrast between the healthy and malignant tissues in DICOM, QHE (Quantum Histogram Equalization) is adopted by using Hadamard-based encoding. Here, image data is mapped into QSV (Quantum State Vector) ψ, the Equation 2 is expressed as,
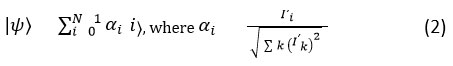
here, QHE enhances local and global contrasts and redistributes the intensity levels allows the high visibility of lung tumors while preserving the tissue structure. In the next stage, the high-frequency noise is removed while retaining the anatomical edges, Wavelet Fusion Enhanced (WFE) method is applied. The raw image is I is decomposed into A called quantum approximation. The detailed coefficients Dh, Dv, Dd (Horizontal, Vertical and Diagonal) is mathematically expressed in Equation 3 is as follows.
![]()
where, thresholding is performed on three coefficients to remove noise without affecting the nodule borders. The final output Iclean will be taken as for denoising and segmentation tasks. After denoising the images, all the features are encoded into QS (Quantum States) for downstream segmentation and classification which is shown in next section. The pixel intensities are represented in superposition state by using the Amplitude Coding method which is expressed in the below Equation 4.
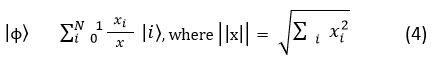
where, the transformation process enables QCL (Quantum Convolutional Layers) to operate efficiently to capture spatial relationships which is not evident in classical convolutional layers.
ROI Segmentation using Quantum Mask-RCNN
The accurate and deep segmentation of lung nodules is crucial for detection and classification tasks. To attain high spatial ROI extraction, a novel Quantum Mask Region-Based CNN (QM-RCNN) combines the classical layers into quantum encoding to enhance boundary recognition, especially in low contrast or ambiguous anatomical regions.
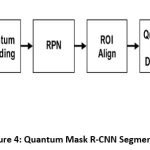 |
Figure 4: Quantum Mask R-CNN Segmentation |
The Mask R-CNN deep segmentation tasks are carried out in five different stages such as,
Input Preparation and Feature Encoding
Region Proposal Network
Quantum-Assisted ROI Alignment
Segmentation and Masking
Output and ROI Extraction
Input Preparation and Feature Encoding
The process begins with the pre-processed CT-DICOM images, which undergo QFT (Quantum Feature Transformation) through the amplitude encoding process. Each image in the pipeline is transformed into HD vector space where each pixel’s intensity level highly contributes to the amplitude of quantum states. This dynamic encoded representation captures the subtle differences in density and texture, which helps discriminate between benign and malignant tumors.
Region Proposal Network (RPN)
RPN predicts bounding boxes for potential nodules. These region proposals are generated based on the anchor boxes with different aspect ratios and scales. Quantum encoding enables the RPN to localize the minute abnormalities accurately, which improves the contrast of faint nodules. The feature maps from this stage serve as input for mask prediction and classification implemented in QEDNN and AFDE frameworks.
Quantum-Assisted ROI Alignment
After the RPN process, each proposed region undergoes ROI alignment, which is enhanced using a quantum-weighted Attention Mechanism. This QWAM technique assigns dynamic weights to each region based on relevance to Q-derived context in order to ensure that nodules with irregular shapes are not misaligned during feature pooling or extraction. The Q-Attention score Qattention for region is computed as,
![]()
where, αi is Q-derived coefficients and fi r is region specific features.
Segmentation and Masking
After the complete process of feature alignment, the aligned features are then passed into quantum-enhanced convolutional decoder which decodes the output as a binary mask for each detected region. Each pixel in the detected mask is assigned a value based on learned spatial patterns which enables the precise boundary detection. The mask loss function Lmask is mathematically expressed in Equation 6 as follows.
![]()
where, Mi and M i is ground truth and predicted mask value of the segmentation process.
Output and ROI Extraction
A set of segmented masks corresponding to individual nodules is extracted as the final output of the QM-RCNN. These masks are fully overlaid on the original DICOM image to extract the ROI patches, and each patch is labeled based on its severity score and passed to the QEDNN classifier for detection and classification. The QM-RCNN segmentation model achieves a Dice Coefficient of 94.6%, which outperforms all existing models in terms of superior boundary conformity and accuracy in anatomical scenarios.
Hybrid QEDNN-AFDE Framework for Tumor Detection and Multi Classification
To overcome the limitations of existing models on precise tumor detection and effective multi-class classification in CT-DICOM images, a hybrid framework is proposed in this research work by integrating Quantum-Enhanced DNN with Adaptive Firefly Differential Evolution optimization mechanism. The novel architecture combines quantum feature encoding techniques, deep pattern extraction, and bio-inspired global optimization methods to enhance diagnostic performance, model interpretability, and computational efficiency. QE-DNN serves as the core classification engine of the system that processes the segmented ROIs extracted via Quantum Mask-RCNN. QE-DNN extends quantum convolutional filters and amplitude coding, enabling the model to capture complex and non-linear features such as tumor density variations, irregularity in edges, and intensity textures from CT-DICOM images. Figure 5 shows the clear architecture model of the proposed QEDNN-AFDE model and hoe it detects and classifies tumor in various stages. The key characteristics of the QE-DNN model are,
Amplitude encoding of inputs – Compress HD pixel values into Quantum probability amplitudes
Quantum Convolutional Layers – Inspired by quantum gates and learns spatial deep features
Multi-Output Classification – Performs multiple tasks simultaneously to attain target classes
Quantum Inspired Activation – Simulates superposition-like behavior using PAF (Parametric Activation Function)
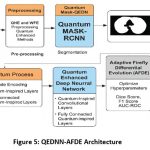 |
Figure 5: QEDNN-AFDE Architecture |
After the process of Quantum amplitude encoding and Convolutional feature mapping which is shown in the previous sections, quantum parametric activation is done to enable smooth transition between activated and inactivated quantum states which is modelled as,
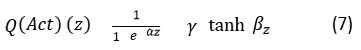
where, α, β, γ are the adaptive parameters which controls the non-linear and simulating super position activation function. In order to activate multi-class total loss function, binary and categorical cross-entropy is combined and expressed in the below equation 8.
![]()
where, γ1 and γ2 balances the tumor detection and classification objectives during the implementation process.
To overcome convergence delays and sub-optimal training in high dimensional deep neural networks, the ADFE mechanism is integrated to optimize the hyperparameter of QE-DNN. AFDE is a bio-inspired optimizer combining firefly exploratory movements with mutation and crossover strategies of Differential Evolution. This dual strategy improves diversity and fine-tuning (Exploration and Exploitation) during training in different iterations/epochs.
In AFDE, firefly movement modeling is the starting phase where fireflies are distributed in the search space, each represents a candidate solution. A firefly moves towards a better (brighter) firefly j based on perceived attractiveness. The firefly attraction movement is expressed mathematically in equation 9.
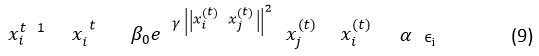
where, β0 denotes initial attractiveness, γ is light absorption coefficient and α is the randomization factor. Here, mutation and crossover take place using DE-based mutation to improve local search. The mutation vector vi is expressed as,
![]()
where, xr1 xr2 xr3 are randomly selected candidate solutions and F denotes the scaling factor. The crossover step generates offspring from parent and mutant vector. The Differential Evolution binomial crossover is represented as,
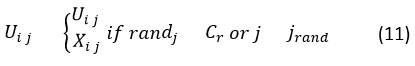
where, Cr is crossover rate and jrand denotes gene selection from the mutant. Finally, fitness evaluation takes place using the fitness function,
![]()
where, offspring with better fitness replace their parents and ensures progressive convergence towards optimal hyper-parameter tuning process. The integrated learning workflow of hybrid QEDNN-AFDE framework operates in iterative training cycles. In every iteration the following steps will be executed and cycled.
ROIs from Q-Mask-CNN will be the input to QEDNN
Amplitude Encoding transforms input to quantum mechanisms
QEDNN processes data using quantum layers
AFDE evaluates QEDNN’s performance and updates the learning rates, dropout rates and weights
Training continues until convergence met based on scores.
The major advantage of using quantum framework is, i) precision learning ii) adaptive tuning, iii) multi-class output capability and iv) generalizability. Also, the model’s ability clearly shows that it is highly fit to work on real-time data to detect and classify the lung tumors as a part of initial screening process in healthcare sector.
Iterative Training and Optimization Strategy
The hybrid QEDNN-AFDE framework is trained using a dynamic, iterative strategy to refine the quantum model’s accuracy, stability, and generalization. This iterative approach integrates a multi-modal bio-inspired feedback-based optimization to adaptively tune hyperparameters, layer-wise configurations, and classification thresholds across multiple learning cycles. In QEDNN, each iteration is an evolutionary step in which the model evolves better to discriminate healthy tissues, malignant nodules, and their severity scores. In every training epoch, iteration-based data feeding is done, where batches of segmented ROI patches are sent as input to QEDNN. Each ROI represents target classes 0, 1, and 2 and includes spatial features such as edge sharpness, texture gradients, and gray-level distribution. At the end of each epoch, AFDE is invoked to evaluate the current QEDNN performance based on the fitness score of candidates. AFDE generates multiple candidate solutions that represent different learning rates and dropout rates. The candidate yields the lowest classification loss, and a high segmentation overlap is selected for the next iteration.
Confusion Matrix and True Value Prediction
A 3×3 confusion matrix is performed to predict the actual values of three target classes: healthy, cancer, and severity score analysis. It is observed that the proposed QEDNN-AFDE model indicates high accuracy and less misclassification during internal training with a 70% ratio of data. During testing, the model maintains high consistency and retains the discriminative power between overlapping categories such as cancer vs severity. During validation, unbiased tuning and consistent precision are tested to predict the false rates and stability of the model to deploy in a real-world clinical decision support system. The model reflects consistent precision during validation. The proposed QEDNN-AFDE provides high model learning consistency and class separation in the training and testing phase with reliable tuning performance. It shows low variance with realistic deployment readiness.
Table 3: Sample iteration values of three target classes
| Confusion Matrix | Correct Healthy Predictions | Correct Cancer Predictions | Correct Severity Predictions |
| Training | 12,100 | 16,750 | 15,540 |
| Testing | 4,100 | 5,700 | 4,980 |
| Validation | 2,050 | 2,825 | 2,495 |
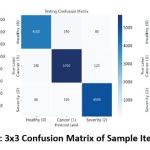 |
Figure 6: 3×3 Confusion Matrix of Sample Iterations |
Quantum Based Bio-Inspired Detection and Classification Process Steps (QEDNN-AFDE)
| 1. Input: CT-DICOM lung cancer image dataset from TCIA repository
2. Begin: Load the dataset DICOM dataset = load(dicomdata.mat) 3. Initial Pre-Processing: Convert all images to grayscale with resolution normalization (512×512 pixels). 4. Perform intensity normalization using min-max scaling for pixel value consistency. 5. Apply QHE to enhance contrast and boundary differentiation. 6. Implement wavelet fusion to suppress background noise while preserving tumor edges. 7. Perform amplitude encoding to map pixel intensity vectors into Qstates. 8. Store Qimages for feature-enhanced processing. 9. Load bounding box annotations and labels Healthy-0, Cancer 1, Severity-2 10. Apply QMASKRCNN to detect and segment lung nodules. 11. Generate segmentation masks to extract QROI from each image. 12. Label each QROI with its corresponding class and assign to training, testing, validation subsets. 13. Initialize QEDNNstructure with predefined structure. 14. Pass QROI through QEDNN using QClayers 15. Extract multi-dimensional spatial and contextual features from QROI. 16. Configure Layeroutput for binary (tumor detection) and multi-class (severity) classification. 17. Initialize AFDEoptimizer for fine-tuning 18. Generate Populationinitial of fireflies representing QEDNN hyperparameter sets. 19. Compute Fitnessscore of each firefly using combined loss 20. Update fireflies using firefly movement, mutation, and DE-based crossover operations. 21. Select Candidatesolution with minimum loss for next QEDNN Trainingiteration 22. Train QEDNN iteratively over multiple epochs with updated parameters. 23. After each epoch, evaluate model using 3×3 confusion matrix on training, test, validation sets. 24. Track performance metrics: Accuracy, Sensitivity, Specificity, F1 Score, Dice Score, AUC-ROC, Log Loss. 25. If performance improvement for 5 epochs or Dice activate early stopping. 26. Store optimal QEDNN-AFDE model parameters for inference. 27. For each test sample, apply segmentation, encode ROI, and classify using trained model. 28. Output classification: Healthy (0), Tumor Detected (1), or Severity Score (2 – mild/moderate/severe). 29. Visualize results with mask overlays, confusion matrices, and metric summaries. 30. End |
Results
The performance evaluation of the proposed QEDNN-AFDE is done using the MATLAB simulation tool to validate the accuracy, generalization, and diagnostic reliability in detecting and classifying the tumors from CT-DICOM images. A 3×3 confusion matrix is performed across multiple iterations to capture three target classes (Healthy (0), Cancer Detected (1), and Severity Level (2)) and truth values. Seven performance evaluation metrics are used, which help reinforce the proposed model’s ability as one of the high-precision diagnostic tools highly suitable for dynamic integration into clinical decision support systems. The equations 13-20 mentioned below are used to calculate the ratio of actual outcomes of each metric across several iterations is shown in the discussions section.
The results of the comparative models are shown and discussed technically in this section. The findings show the robust detection and classification of the QEDNN-AFDE model against existing models such as SVM-WSS7, GCPSO-PNN8, 3D-DLCNN9, and ECNNDE-BCE10. The MATLAB graph of all the metrics shows the output values where the X-axis denotes models, and the Y-axis denotes percentages. Figure 7-12 indicates the performance evaluation metrics and discussions on how the model reflects effective detection and classification of lung tumors under various iterations. The new model outperforms all the existing models across all metrics and iterations. Various epochs with different batch sizes are carried out to get desired output values.
where, MCC denotes Matthews Correlation Coefficient and the true values are derived by the equation, T1 = (TPR × TNR – FPR × FNR) T2 = (TPR + FPR), T3 = (TPR + FNR), T4 = (TNR + FPR), and T5 = (TNR + FNR).
Sensitivity and Specificity
Measures the model’s ability to accurately detect actual cancer and non-cancerous cases. High sensitivity ensures that malignant cases are not missed, and high specificity ensures that no healthy individuals are wrongly diagnosed.
Accuracy
The overall correctness of predictions by calculating the ratio of positive and negative against total samples. This metric validates the target classes and their outcomes.
AUC-ROC
The metric quantifies the model’s ability to distinguish the TPR, FPR, and FNR between target classes. Higher AUC indicates better discriminative performance in robustly separating the classes.
Log Loss
The confidence levels are measured, and the predicted probability distribution is checked to see how close it is to the actual label. Lower log loss ensures that the outputs are trustworthy for clinical use.
Dice Coefficient
Spatial overlap metric, especially for segmentation tasks, compares predicted region and ground truth and assesses how accurately the lung nodules are localized.
F1 Score
It balances the FPR and FNR and offers more accuracy in imbalanced datasets with minimal trade-offs.
Discussions
Sensitivity and Specificity Comparative Analysis
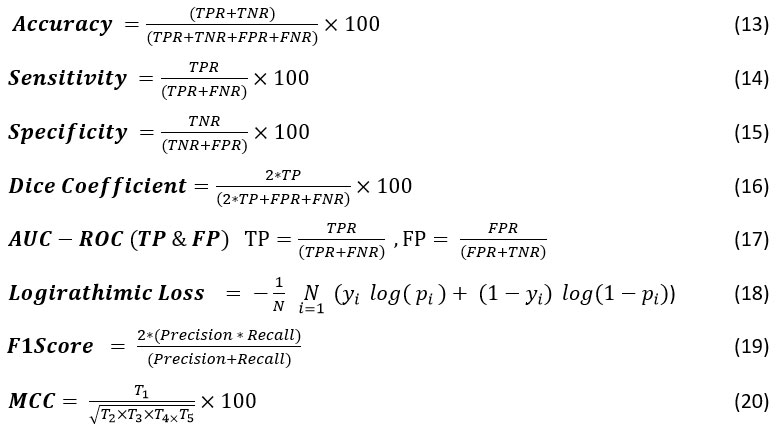
The sensitivity and specificity analysis of the proposed quantum-based bio-inspired QEDNN-AFDE model shown in Figure 7, achieved an exceptional diagnostic performance of 95.2% and 95.8%, reflecting the strength in detecting actual tumor cases by excluding healthy instances. The quantum pre-processed masks ensure accurate region extraction in this model, improving classification boundaries. The quantum-enhanced convolutional layers preserve subtle diagnostic features, and AFDE fine-tunes the hyperparameters to optimize recall and precision simultaneously. QEDNN-AFDE consistently discriminates against cancer and health across varying nodule shapes and densities, helping the model be suitable for deployment in real-time scenarios for cancer screening and severity score analysis in the healthcare sector.
Table 4: Sensitivity and Specificity Analysis
| Metrics / Schemes | SVM-WSS7 | GCPSO-PNN8 | 3D-DLCNN9 | ECNN-BCE10 | QEDNN-AFDE (Proposed) |
| Sensitivity | 76.52 | 81.78 | 86.35 | 93.81 | 95.20 |
| Specificity | 78.65 | 83.40 | 88.48 | 94.52 | 95.80 |
 |
Figure 7: Sensitivity and Specificity Graph |
Accuracy Comparative Analysis
Figure 8 shows the overall accuracy of the proposed QEDNN-AFDE model, which attains 96.4%, confirming its ability to identify and classify the target classes such as cancer, healthy, and severity scores with the help of DICOM image datasets. QEDNN overcomes the limitations of existing models and allows deep extraction and multi-level spatial feature extraction from CT-DICOM images. The AFDE fine-tunes the learning parameters dynamically to ensure deep convergence towards optimal weights across various iterations, which helps to minimize FPR and FNR. The high accuracy of this QEDNN-AFDE reflects the model’s suitability in multi-class environments, which improves the generalization without compromising the diagnostic precision, making the system trustworthy for dynamic integration in clinical decision-support pipelines.
Table 5: Accuracy Rate Analysis
| Metrics / Schemes | SVM-WSS7 | GCPSO-PNN8 | 3D-DLCNN9 | ECNN-BCE10 | QEDNN-AFDE (Proposed) |
| Accuracy (It-1) | 71.26 | 78.48 | 84.52 | 92.45 | 94.91 |
| Accuracy (It-N) | 73.18 | 80.08 | 87.68 | 94.73 | 96.40 |
 |
Figure 8: Accuracy Analysis Graph |
AUC-ROC Analysis
The performance analysis of the AUC-ROC metric is showcased in Figure 9. This metric serves as a robust discriminative evaluation across all target classes. The QEDNN-AFDE exhibits 0.98 AUC, indicating the ability to classify healthy, cancerous, and severity-score lung images. The higher AUC denotes that the hybrid architecture is tuned ideally based on thresholds. The quantum encoding mechanism allows the preservation of high dimensional texture cues while AFDE adjusts the classification boundaries for each class. This dual method resulted in distinct score distributions, making them more evident in ROC space. The multi-class ROC analysis consistently shows high TPR and FPR across all categories, which is scalable for diverse lung pathology datasets. The results of the proposed QEDNN-AFDE model are compared against existing models SVM-WSS7, GCPSO-PNN8, 3D-DLCNN9, and ECNNDE-BCE10. The promising results show that QEDNN-AFDE outperforms the existing models with high discrimination power in accurate classification and smooth diagnosis capability.
Table 6: AUC-ROC Analysis
| Metrics / Schemes | SVM-WSS7 | GCPSO-PNN8 | 3D-DLCNN9 | ECNN-BCE10 | QEDNN-AFDE (Proposed) |
| TPR | 0.62 | 0.70 | 0.79 | 0.93 | 0.95 |
| FNR | 0.39 | 0.31 | 0.23 | 0.11 | 0.05 |
 |
Figure 9: AUC-ROC Analysis Curve |
Dice Coefficient Analysis
The comparative result analysis of the dice coefficient metric is shown in Figure 10. This metric is a spatial overlap to evaluate the segmentation quality. The proposed QEDNN-AFDE achieved a dice score of 94.6%, demonstrating its ability to localize and segment the lung nodules in CT-DICOM images accurately. This was achieved by quantum Mask-RCNN, which enhanced the ROI boundaries through quantum-assisted attention mechanisms. QHE and WEF ensure edge preservation to improve ROI mask clarity and alignment with actual nodular structures. QEDNN-AFDE identifies irregular shapes, small tumors, and overlapping anatomical structures in the DICOM dataset. The model minimized FNR within nodules and reduced false positive markings outside the target areas. The consistent results with high-overlap segmentation output are validated across multiple epochs and iterations. This model helps in pre-treatment planning, surgical mapping, and tumor progression analysis.
Table 7: Dice Coefficient Analysis
| Metrics / Schemes | SVM-WSS7 | GCPSO-PNN8 | 3D-DLCNN9 | ECNN-BCE10 | QEDNN-AFDE (Proposed) |
| Dice Score (It-1) | 77.19 | 83.17 | 85.45 | 90.78 | 92.81% |
| Dice Score (It-N) | 79.80 | 85.16 | 87.76 | 92.44 | 94.60% |
 |
Figure 10: Dice Coefficient Graph |
Logarithmic Log Analysis
A comparative analysis of the logarithmic loss of different models is shown in Figure 11. It evaluates the confidence of probability-based predictions, where lower values indicate fewer errors. The proposed QEDNN-AFDE achieved low log loss due to a robust probabilistic learning design. As QDNN uses the parameter shift rule, the prediction probabilities are maximized in all iterations. This strategy is mainly used in borderline cases, where overconfident misclassification scores are standard in deep neural networks. The low log loss validates the ability of the proposed quantum-based model to generate reliable confidence scores, which are essential for clinical decision-making, especially in automated reported systems to assess diagnostic risk.
Table 8: Log Loss Score Analysis
| Metrics / Schemes | SVM-WSS7 | GCPSO-PNN8 | 3D-DLCNN9 | ECNN-BCE10 | QEDNN-AFDE (Proposed) |
| LLS (It-1) | 0.47 | 0.35 | 0.26 | 0.10 | 0.08 |
| LLS (It-N) | 0.37 | 0.28 | 0.19 | 0.04 | 0.02 |
 |
Figure 11: Log Loss Graph |
F1-Score Comparative Analysis
Figure 12 shows the harmonic mean analysis called F1 Score of the proposed QEDNN-AFDE to evaluate underclass imbalance, which is common in clinical datasets. The proposed work shows a remarkable 95.2% F1 score, which signifies its balanced performance in detecting TPR and avoiding False rates. The hybrid quantum convolutional layers extracted spatial-temporal features that preserved essential tumor patterns in low-contrast regions. This dynamic learning strategy allows the proposed QEDNN-AFDE model to maintain high performance and consistency across all three target classes. A high F1 score reflects the diagnostic fidelity of the quantum model and highlights the capability to generalize input conditions and imaging variations across many patients. This will act as a pre-requisite for AI-based real-time deployment in radiology workflows.
Table 9: Analysis of F-Score
| Metrics / Schemes | SVM-WSS7 | GCPSO-PNN8 | 3D-DLCNN9 | ECNN-BCE10 | QEDNN-AFDE (Proposed) |
| F1 Score (It-1) | 75.35 | 80.68 | 86.55 | 91.30 | 93.74 |
| F1 Score (It-N) | 78.18 | 82.74 | 88.76 | 93.42 | 95.20 |
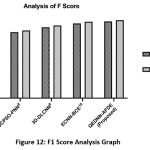 |
Figure 12: F1 Score Analysis Graph |
Merits and Demerits of the Proposed Technique
The proposed QEDNN-AFDE model significantly outperforms baseline models such as SVM-WSS, GCPSO-PNN, 3D-DLCNN, and ECNN-BCE by utilizing the quantum-inspired feature encoding and adaptive swarm-based hyperparameter optimization. The key merits include enhanced convergence speed, superior sensitivity and specificity, and improved generalization across varying clinical CT-DICOM large-scale datasets. Unlike baseline optimization models, QEDNN-AFDE effectively minimizes overfitting through dynamic search capabilities. However, the computational complexity of QEDNN and the resource overhead introduced by AFDE may pose challenges in low-resource environments. Additionally, integration of QAE requires more careful calibration compared to conventional CNN layers. Despite these constraints, the method demonstrates considerable promise in early lung cancer detection which offers interpretability, scalability, and accuracy gains over prevailing techniques.
Conclusion
This proposed QEDNN-AFDE research work presents a dynamic integration of quantum deep learning and bio-inspired optimization to address the limitations of lung tumor detection and severity analysis from CT-DICOM images. QE-DNN and Q-CNN stand robust foundation of the model, successfully operates synergistically to detect healthy and cancerous tissues with tumor severity scores. Advanced pre-processing techniques such as histogram equalization and wavelet fusion ensure structural clarity analysis across complex features of DICOM. The bio-inspired AFDE algorithm improves the hyperparameter significantly, leading to a well-regularized and high-performing model across diverse patient image variations. The outperforming results with 96.4% accuracy, 95.2% sensitivity, 95.8% specificity, 95.2% F1 score, 94.6% dice coefficient, 0.02 Log Loss and AUC-ROC with 0.95 TPR, and 0.05 FNR highlights the model’s potential for early and accurate diagnosis, prognosis planning and decision support in oncology workflows. The study elevates the diagnostic capabilities of dep-learning integration in radiology and demonstrates the feasibility of a quantum-inspired framework for real-world medical applications. By merging advanced computational methods with biologically motivated learning strategies, QEDNN-AFDE establishes a new benchmark in precision image analysis.
Though the model achieves outstanding performance, the implementation is limited to quantum simulators rather than actual quantum hardware. Also, real-time deployment in clinical environments requires validation under imaging protocols and multi-institutional datasets to ensure the model’s robustness.
Acknowledgement
The authors would like to express their sincere gratitude to their respective institutions, PKR Arts College for Women, Gobichettipalayam, Tamil Nadu and PPG College of Arts & Science, Affiliated to Bharathiar University, Coimbatore, Tamilnadu, India for their support throughout this research.
Funding Sources
The author(s) received no financial support for the research, authorship, and/or publication of this article.
Conflict of interest
The authors do not have any conflict of interest.
Data Availability Statement
This statement does not apply to this article. Simulation data will be provided on request.
Ethics Statement
This research did not involve human participants, animal subjects, or any material that requires ethical approval.
Informed Consent Statement
This study did not involve human participants, and therefore, informed consent was not required.
Clinical Trial Registration
This research does not involve any clinical trials, only software simulations are carried out.
Author Contribution
- Kalaivani Devaraj has collected the data and identified problem statement and objectives as an initial phase. Also, the complete methodology part is done by her with all implementation using simulation tools and played a key role in refining the manuscript as a corresponding author.
- Dheepa Ganapathy has done a thorough paper evaluation, language check, supervision and guidance for implementation as a co-author and supervisor and made all the editorial checks, paper revision etc. Both the authors have given their approval for its publication.
References
- Zia UrRehman, Yan Qiang, Rukhma Aftab & Juanjuan Zhao. Effective lung nodule detection using deep CNN with dual attention mechanisms. Scientific Reports. 2024;14(1). DOI: https://doi.org/10.1038/s41598-024-51833-x
CrossRef - Martis JE, M S S, R B, Mutawa AM, Murugappan M. Novel Hybrid Quantum Architecture-Based Lung Cancer Detection Using Chest Radiograph and Computerized Tomography Images. 2024; 11(8):799. https://doi.org/10.3390/bioengineering11080799
CrossRef - R. N, Chandra S. S. V. ExtRanFS: An Automated Lung Cancer Malignancy Detection System Using Extremely Randomized Feature Selector. Diagnostics. 2023; 13(13):2206. https://doi.org/10.3390/diagnostics13132206
CrossRef - Wahab Sait AR. Lung Cancer Detection Model Using Deep Learning Technique. Applied Sciences. 2023; 13(22):12510. https://doi.org/10.3390/app132212510
CrossRef - Klangbunrueang R, Pookduang P, Chansanam W, Lunrasri T. AI-Powered Lung Cancer Detection: Assessing VGG16 and CNN Architectures for CT Scan Image Classification. 2025; 12(1):18. https://doi.org/10.3390/informatics12010018
CrossRef - S Nithyanandh and V Jaiganesh. Quality of service enabled intelligent water drop algorithm based routing protocol for dynamic link failure detection in wireless sensor network. Indian Journal of Science and Technology. 2020;13(16):1641-1647. https://doi.org/10.17485/ijst/v13i16.19
CrossRef - Chaturvedi P, Jhamb A, Vanani M, Nemade V. Prediction and classification of lung cancer using machine learning techniques. IOP Conference Series Materials Science and Engineering. 2021;1099(1):012059. https://doi.org/10.1088/1757-899x/1099/1/012059
CrossRef - Jagadeesh K, Rajendran A. Improved model for Genetic Algorithm-Based Accurate lung cancer segmentation and classification. Computer Systems Science and Engineering. 2022;45(2):2017-2032. https://doi.org/10.32604/csse.2023.029169
CrossRef - Eldho KJ, Nithyanandh S. Lung Cancer Detection and Severity Analysis with a 3D Deep Learning CNN Model Using CT-DICOM Clinical Dataset. Indian Journal of Science and Technology. 2024;17(10):899-910. https://doi.org/10.17485/ijst/v17i10.3085
CrossRef - Kalaivani D, Dheepa G. Deep Learning Enhanced CNN with Bio-Inspired Techniques and BCE For Effective Lung Nodules Detection & Classification For Accurate Diagnosis. Indian Journal of Science and Technology. 2024;17(37):3851-3864. https://doi.org/10.17485/ijst/v17i37.2649
CrossRef - Wei L, Liu H, Xu J, et al. Quantum machine learning in medical image analysis: A survey. Neurocomputing. 2023;525:42-53. https://doi.org/10.1016/j.neucom.2023.01.049
CrossRef - Nithyanandh S, Omprakash S, Megala D, Karthikeyan MP. Energy Aware Adaptive Sleep Scheduling and Secured Data Transmission Protocol to enhance QoS in IoT Networks using Improvised Firefly Bio-Inspired Algorithm (EAP-IFBA). Indian Journal of Science and Technology. 2023;16(34):2753-2766. https://doi.org/10.17485/ijst/v16i34.1706
CrossRef - Ahmad I, Alqurashi F. Early cancer detection using deep learning and medical imaging: A survey. Critical Reviews in Oncology/Hematology. 2024;204:104528. https://doi.org/10.1016/j.critrevonc.2024.104528
CrossRef - Bharathi PS, Shalini C. Advanced hybrid attention-based deep learning network with heuristic algorithm for adaptive CT and PET image fusion in lung cancer detection. Medical Engineering & Physics. 2024;126:104138. https://doi.org/10.1016/j.medengphy.2024.104138
CrossRef - S Nithyanandh and V Jaiganesh. Dynamic Link Failure Detection using Robust Virus Swarm Routing Protocol in Wireless Sensor Network. International Journal of Recent Technology and Engineering (IJRTE). 2019;8(2):1574-1579. https://doi.org/10.35940/ijrte.b2271.078219
CrossRef - Elhassan SM, Darwish SM, Elkaffas SM. An enhanced lung cancer detection approach using Dual-Model Deep Learning technique. Computer Modeling in Engineering & Sciences. 2024;0(0):1-10. https://doi.org/10.32604/cmes.2024.058770
CrossRef - Choudhury AR, Rautray J, Mishra P, Kandpal M, Dalai SS. Deep Learning Based Automated Lung Cancer Detection from CT scan Leveraging Transfer Learning. Procedia Computer Science. 2025;258:2748-2759. https://doi.org/10.1016/j.procs.2025.04.535
CrossRef - Nithyanandh S, Jaiganesh V. Reconnaissance Artificial Bee Colony Routing Protocol to Detect Dynamic Link Failure in Wireless Sensor Network. International Journal of Scientific & Technology Research. 2019; 10(10):3244–3251. https://www.ijstr.org/final-print/oct2019/Reconnaissance-Artificial-Bee-Colony-Routing-Protocol-To-Detect-Dynamic-Link-Failure-In-Wireless-Sensor-Network.pdf
- Sharma D, Choubey DK, Thakur K. Lung and Colon Cancer Detection using Deep Learning Techniques. Procedia Computer Science. 2025;258:4136-4146. https://doi.org/10.1016/j.procs.2025.04.664
CrossRef - Arularasan R, Balaji D, Garugu S, Jallepalli V R, Nithyanandh S, Singaram G. Enhancing Sign Language Recognition for Hearing-Impaired Individuals Using Deep Learning. 2024 International Conference on Data Science and Network Security, Tiptur, India. 2024; 10690989:1-6.
https://doi.org/10.1109/icdsns62112.2024.10690989
CrossRef - Devi PA, Megala D, Paviyasre N, Nithyanandh S. Robust AI Based Bio Inspired Protocol using GANs for Secure and Efficient Data Transmission in IoT to Minimize Data Loss. Indian Journal of Science and Technology. 2024;17(35):3609-3622. https://doi.org/10.17485/ijst/v17i35.2342
CrossRef - Abe AA, Nyathi M, Okunade AA, Pilloy W, Kgole B, Nyakale N. A Robust Deep Learning Algorithm for Lung Cancer Detection from Computed Tomography Images. Intelligence-Based Medicine. January 2025:100203. https://doi.org/10.1016/j.ibmed.2025.100203
CrossRef - Crasta LJ, Neema R, Pais AR. A novel Deep Learning architecture for lung cancer detection and diagnosis from Computed Tomography image analysis. Healthcare Analytics. 2024;5:100316. https://doi.org/10.1016/j.health.2024.100316
CrossRef - Prabhu TS, Nithyanandh S, Eldho KJ, Karthikeyan B, Vasanthi V. Securing Next Generation 6G Wireless Networks Through Intelligent Bio-Inspired Routing with Energy Optimization for Enhanced Authentication. Indian Journal of Science and Technology. 2025;18(23):1882-1895. https://doi.org/10.17485/ijst/v18i23.850
CrossRef - Hroub NA, Alsannaa AN, Alowaifeer M, Alfarraj M, Okafor E. Explainable deep learning diagnostic system for prediction of lung disease from medical images. Computers in Biology and Medicine. 2024;170:108012. https://doi.org/10.1016/j.compbiomed.2024.108012
CrossRef - Selvam N, Joy EK. Plant Leaf Disease Detection with Multivariable Feature Selection Using Deep Learning AEN and Mask R-CNN in PLANT-DOC Data. Biosciences Biotechnology Research Asia. 2024;21(4):1649-1663. https://doi.org/10.13005/bbra/3333
CrossRef - Nithyanandh S. Object Detection & Analysis with Deep CNN and Yolov8 in Soft Computing Frameworks. International Journal of Soft Computing and Engineering. 2025;14(6):19-27. https://doi.org/10.35940/ijsce.e3653.14060125
CrossRef - Gao Z, Guo Y, Wang G, et al. Robust deep learning from incomplete annotation for accurate lung nodule detection. Computers in Biology and Medicine. 2024;173:108361. https://doi.org/10.1016/j.compbiomed.2024.108361
CrossRef - Li L, Mei Z, Li Y, Yu Y, Liu M. A dual data stream hybrid neural network for classifying pathological images of lung adenocarcinoma. Computers in Biology and Medicine. 2024;175:108519. https://doi.org/10.1016/j.compbiomed.2024.108519
CrossRef - Kwon H, Ko S, Ha K, Lee JK, Choi Y. Assessing the predictive ability of computational epitope prediction methods on Fel d 1 and other allergens. PLoS ONE. 2024;19(8):e0306254. https://doi.org/10.1371/journal.pone.0306254
CrossRef - Li P, Wang S, Li T, Lu J, HuangFu Y, & Wang D. A Large-Scale CT and PET/CT Dataset for Lung Cancer Diagnosis (Lung-PET-CT-Dx). The Cancer Imaging Archive, https://doi.org/10.7937/TCIA.2020.NNC2-0461 (Dataset Collected Source)

This work is licensed under a Creative Commons Attribution 4.0 International License.





