Manuscript accepted on : 19-06-2025
Published online on: 27-06-2025
Plagiarism Check: Yes
Reviewed by: Dr. Balram Ji Omar
Second Review by: Dr. Aisha Belal
Final Approval by: Dr. Ali Elshafei
Efflux Pump Inhibitors and their Role in Combating Bacterial Drug Resistance
Benjamin Isaac Thomson1* and Shainaba A Saadhali2
and Shainaba A Saadhali2
1Department of Biotechnology, School of Bioengineering, SRM Institute of Science and Technology, Kattankulathur, Tamil Nadu, India
2Department of Bacteriology, ICMR- National Institute for Research in Tuberculosis, Chetpet, Chennai, India
Corresponding Author E-mail:bteben02@gmail.com
ABSTRACT: The use of efflux pump inhibitors (EPIs) as adjuvant therapy to combat bacterial resistance to several antibiotics has been a proven method to fight antimicrobial resistance. Antibiotic- expelling efflux pumps, which are found in bacterial cell membranes, play a major role in the development of multidrug resistance. Therefore, EPIs, which are meant to inhibit these pumps, have the potential to make the current antibiotic arsenal more effective. Here we classify efflux pumps, analyse the many complexities of EPI processes, and emphasize the range of sources and applications. This academic review thus highlights the functioning of efflux pumps, the critical role that efflux pump inhibitors play and advocates for more research into efflux pump inhibitors.
KEYWORDS: Antimicrobial resistance; Efflux pump; Efflux pump inhibitors; Multi-drug resistance; Mycobacterium tuberculosis
| Copy the following to cite this article: Thomson B. I, Saadhali S. A. Efflux Pump Inhibitors and their Role in Combating Bacterial Drug Resistance. Biotech Res Asia 2025;22(3). |
| Copy the following to cite this URL: Thomson B. I, Saadhali S. A. Efflux Pump Inhibitors and their Role in Combating Bacterial Drug Resistance. Biotech Res Asia 2025;22(3). Available from: https://bit.ly/3Tq3ahG |
Introduction
The damage to the efficacy of several drugs resulting from bacteria resistant is a significant obstacle to traditional healthcare, affecting the efficacy of our powerful antimicrobial treatments.1 Although there are multiple modes of drug resistance, efflux pumps specifically leave no margin for antibiotics to have a therapeutic effect within the microorganism.2
This obstacle which causes our present antibiotics to become obsolete has brought the focus on the need for exploration into innovative approaches to combat bacterial resistance. The use of efflux pump inhibitors [EPIs] in adjuvant therapy to increase the effectiveness of current antibiotics is one such tactic that has been shown to produce effective results.3,4
Antibiotic-specific efflux pumps, bedded within the bacterial cell membranes, play a vital part in the development of multi-drug resistance by extensively expelling antibiotics from bacterial cells.5–7 These efflux pumps, when overexpressed or hyperactive due to evolution or environmental factors, contribute significantly to the survival and continuity of antibiotic- resistant microorganism.8
Researchers have focused on utilizing compounds known to have efflux pump-inhibiting properties which are able to restore antibiotic sensitivity of bacteria by blocking these efflux pumps.9 The logic behind EPI is that they can enhance the efficiency of antibiotics by preventing their exit from bacterial cells, thereby increasing the intracellular activity of the drug at the target point.
This approach has the potential to bring back the clinical viability of current antibiotics and increase the lifetime of our obsolete antibiotics. The field of EPIs has witnessed an increase in research in recent times, with a growing body of literature pressing their eventuality as adjuvants in antibiotic remedy. These inhibitors not only offer a means to combat resistance but also give a platform for the development of new remedial strategies that could revise the treatment of bacterial infections.10
In this review, we take a glimpse into the world of EPIs and how they are used in adjuvant treatment of various health complications. The mechanisms of antibiotic resistance intermediated by efflux pumps, the strategies employed to identify and develop EPIs, and their part in enhancing the efficacy of various antibiotics are also explored.
Mechanism of Efflux Pumps
Specialized membrane proteins called efflux pumps are present in bacterial cells, and their main job is to drive out harmful substances – including antibiotics – from the cell, making the drug ineffective. Here, we will provide a detailed explanation of the mechanism of efflux pumps including the various types of efflux pumps.11,12
The efflux pump is thought to be a complex protein complex that plays a role in both drug resistance and bacterial pathogenicity.13 Efflux pumps contain three main components:
Transmembrane transporter – Allows transfer of substances across the membrane
Membrane fusion protein – Acts as communicator between outer membrane component and transporter
Outer membrane factor – Helps in the final extrusion of the substance
Another important factor involved in efflux mechanism is adenosine triphosphate [ATP] which provides the efflux pump with the necessary energy required for the process. For example, the NorA efflux pump identified in S. aureus utilizes energy from the hydrolysis of ATP to extrude fluoroquinolone, thus lessening its effects.14
The following are the steps that comprise the efflux pump mechanism:
Binding
The unique chemical form of the efflux pump decides the different kinds of chemicals that it recognizes. Binding sites that correspond with the structure of the drug specific efflux pump form a major part of an EPI’s architecture. The harmful substance [antibiotics or other toxic substances synthesized within the bacteria] binds to the efflux pump due to chemical reactions involving Van der Waals forces, electrostatic interactions, and hydrogen bonds. Additionally, they have the ability to attach to the efflux pump site by altering the pump protein’s shape.
Activation
Efflux pumps actively move the bound material against its concentration gradient by harnessing cellular energy. The toxic chemical binds to the pump, causing it to hydrolyze ATP and alter its conformation. The material can now cross the cell membrane and be released from the cell as a result of this alteration.
Transport
Active transport is used by the efflux pump to remove undesirable materials from the bacterial cell. The efflux pump is dormant until an antagonist attaches itself to it. This initiates the process of expelling the material by using ATP as energy. The energy required for the ejection is provided by the hydrolysis of ATP, which converts ATP into inorganic phosphate and ADP. Efflux pumps continuously identify toxins in its cytoplasmic environment to prevent the accumulation of the substance. Bacteria with highly effective and selective efflux pumps can pump out antibiotics which renders the antibiotics ineffective.
Ejection
And finally, the harmful substance is ejected out15. The efflux pump returns to its original state following the ejection, ready for a subsequent round of ejection if necessary. Thus, this clever mechanism allows bacteria to thrive in the face of hardship.16,17
Diverse bacteria use different versions of the pump, each with its own set of structural features and functions. This diversity justifies the need to categorize efflux pumps into several types.
Efflux pumps can be generically divided into five families according to different structural properties and sequencing. Every family is further subdivided into different kinds of efflux pumps, providing a wide range of options that correspond with the diverse requirements of bacterial life.
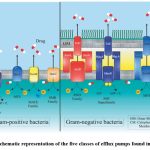 |
Figure 1: Schematic representation of the five classes of efflux pumps found in bacteria18. |
Major Facilitator Superfamily [MFS]
The MFS family is one of the largest, with pumps that have a wide range of substrate specificity. MFS pumps are one-component systems, which means they rely solely on a membrane transporter. The NorA pump in Staphylococcus aureus is a good example. Sugars, ions, amino acids, peptides, drugs, and other small molecules are among the substrates transported across biological membranes by MFS proteins. They are required for a variety of physiological processes such as nutrient uptake, drug resistance, and solute transport.19
MFS proteins cross the lipid bilayer by means of 12–14 alpha-helical transmembrane regions. The transmembrane segments constitute the central domain of the protein, facilitating the passage of substrates through the membrane. Cytoplasmic loops that link the transmembrane segments are another feature of MFS proteins. Functional domains or motifs involved in substrate binding, transport control, or connections with cellular signaling pathways are frequently found in these loops.19,20
ATP-Binding Cassette [ABC] family
ATP serves as the direct energy source for ABC efflux pumps. These are complex systems that function as a unit and include transporter proteins, auxiliary proteins, and ATP-binding proteins21. All living things contain members of one of the biggest and most prevalent protein families: the ATP-Binding Cassette [ABC] transporters. These transporters are involved in the active transport of a wide variety of substrates, such as medicines, carbohydrates, ions, amino acids, lipids, and peptides, across cellular membranes.22 The regulation of intracellular concentrations of ions and metabolites, cellular detoxification, and food uptake are just a few of the physiological processes in which ABC transporters are essential.
ABC transporters create a conduit through the lipid bilayer by combining several transmembrane domains. The recognition and transportation of substrates across the membrane are carried out by these transmembrane domains [TMDs]. ABC transporters also have Cytoplasmic Nucleotide-Binding Domains [NBDs] which are also capable of binding and hydrolyzing ATP. The energy needed for substrate transport is provided by ATP binding and hydrolysis. Most NBDs are common to all ABC transporters. Periplasmic Binding Proteins are found in some ABC transport systems, particularly in bacteria. These proteins carry substrates to the transmembrane domains by capturing them from the surrounding environment.23
Small Multidrug Resistance [SMR] family
Bacteria contain SMR pumps, which are capable of handling a large variety of tiny, hydrophobic cationic compounds. There are just about 100–120 amino acids in these little pumps. Drugs and antimicrobial agents are among the harmful substances that SMR proteins help flush out of the cell and into the extracellular environment. SMR proteins do not require ATP hydrolysis in order to function, in contrast to ATP-binding cassette [ABC] transporters. Rather, they derive energy for drug efflux from the proton motive force [PMF] across the cell membrane.
SMR proteins typically consist of four transmembrane helices. These helices form a channel that spans the lipid bilayer of the cell membrane. SMR proteins also have short cytosolic loops connecting the transmembrane helices. These loops are involved in substrate recognition and transport.24
Resistance-Nodulation-Division [RND] family
RND pumps have a high level of antibiotic resistance, particularly in Gram-negative bacteria. Because of their unique periplasmic funnel. These tripartite systems can extrude a wide variety of substrates. RND transporters are involved in the efflux of a wide variety of substrates from the bacterial cell to the extracellular environment, including antibiotics, heavy metals, detergents, and other toxic compounds. These transporters are important in the context of antibiotic resistance because they play a critical role in bacterial resistance to various antimicrobial agents.25,26
Usually, RND proteins have between ten and twelve transmembrane helices. The helices in Gram-negative bacteria create a structure akin to a tunnel that connects their outer and inner membranes, facilitating the movement of substrates from the cytoplasm to the beyond. The periplasmic domain of RND transporters is situated in the space between the inner and outer membranes. Substances from the periplasmic space are taken up by this region and sent to the transmembrane channel for efflux. RND transporters bind to proteins in the outer membrane, including OMF, to form complexes that open up a channel in the membrane. Substrates can be discharged into extracellular space through this route.27
Multi-Antimicrobial Extrusion [MATE] family
MATE pumps often use a gradient of Na+ or H+ ions and release a wider variety of substrates, such as medications and metabolites. MATE transporters play a crucial role in the efflux of various substrates, including antibiotics, organic ions, and other toxic compounds, from the cell to the extracellular environment. Like other efflux transporter families, MATE transporters are involved in multidrug resistance and contribute to the ability of microorganisms to survive in the presence of antimicrobial agents.28
Twelve transmembrane helices on average make up MATE transporters, which create a channel that spans the lipid bilayer. These helices are in charge of transporting and identifying substrates. The transmembrane helices of MATE transporters are connected by cytosolic loops. Conserved motifs involved in substrate binding and transport are frequently found in these loops.
It’s crucial to understand that the aforementioned families are not strict classes; rather, because of the bacterial efflux pumps’ varied functionality and quick evolution, overlapping instances may occur. Additionally, in order to ensure survival in a variety of environments, the same bacterium may employ pumps from distinct families.11
Efflux Pump Inhibitor Activity in Various Scenarios
EPI can provide direct or indirect therapeutic benefits in a variety of techniques and applications. Bacterial cells use efflux pumps as defense mechanisms to eliminate “noxious” substances from their interior which also include antibiotics.29,30 A research team utilized whole-cell assays to identify efflux pump inhibitors [EPIs] and discovered three multidrug resistance efflux pumps [MexAB-OprM, MexCD-OprJ, and MexEF-OprN] that play a role in fluoroquinolone resistance in clinical strains of Pseudomonas aeruginosa. Additionally, they found that AcrAB-ToIC, which is a close relative of the Escherichia coli efflux pump, along with the three P. aeruginosa. Mex efflux pumps, can be inhibited by the compound “MC-207,110”. This is a property of broad-spectrum EPIs wherein the same compound can be used to inhibit multiple efflux pumps that are similar in structure and function8. When MC-207,110 was utilized, P. aeruginosa’s inherent resistance to fluoroquinolones was greatly decreased.31
Enterobacter aerogenes, one of the most often identified nosocomial infections, exhibits heightened drug resistance which are linked to altered membrane permeability. The main causes of resistance to quinolone medications are changes in the target and active efflux. The impact of several pyridoquinoline compounds on the accumulation of fluoroquinolones in resistant strains that overexpress MarA activator was documented by researchers. Studies on the energy-dependent quinolone efflux indicate that the most potent derivatives investigated probably block the resistance mechanism by posing a competition for substrate with the pump that extrudes intracellular norfloxacin.32 Another study looked at the efflux mechanism in S. aureus and evaluated metformin, domperidone, diclofenac, and glyceryl trinitrate as possible EPIs that could be used in conjunction with antibiotics to treat S. aureus-caused topical infections. By lowering the antibiotic’s minimum inhibitory concentrations and dramatically lowering the relative expression of efflux genes, the efflux assay demonstrated that the tested medications may have efflux inhibitory effects.33,34
Compounds derived from plant extracts are also included in the various categories of EPIs35. These compounds have more advantages than regular EPI since they have far fewer side effects and are highly specific. In particular, terpenes, which are a type of natural product with the chemical formula [C5H8]n for n equal to 2, are significantly more effective at inhibiting EP in both Gram-positive and Gram-negative bacteria. However, Gram-positive strains show a notably higher frequency of inhibition.36,37 Medicinal plants are the most promising natural source of EPIs since they contain a diverse range of secondary metabolites with varying pharmacological properties. Several studies on extracts from medicinal plants have found putative components that inhibit efflux pumps in both Gram-positive and Gram-negative bacteria.38,39 These chemicals may also be able to restore antibiotic efficacy by allowing drugs to reach high enough concentrations inside bacteria to have a bactericidal impact. Many isolated compounds with strong EPI action indicate that some families may be good as EPI providers. Apocynaceae, Berberidaceae, Cucurbitaceae, Convolvulaceae, Fabaceae, Lamiaceae, and Zingiberaceae are a few examples.40
Mycobacterium Tuberculosis as an Efflux Pump Inhibitor Target
The most common, fatal disease worldwide in 2019 was tuberculosis [TB], an infectious disease brought on by the bacteria M. tuberculosis.41,42 Naturally, due to M. tuberculosis’s veracity, several MDR strains of M. tuberculosis have been identified and isolated. The majority of strains employ efflux pumps to maintain multi-drug resistance. Rv1218c is an efflux pump that has been shown to be a viable target for treating clinical isolates of M. tuberculosis that exhibit resistance to multiple drugs. As a result, the inhibition of Rv1218c was evaluated using eight efflux pump inhibitors [EPIs] chosen through silico methods. These compounds were subjected to the ethidium bromide-DNA binding assay, the minimum inhibitory concentration [MIC] assay, the checkerboard drug combination assay, and the in vitro and ex vivo cytotoxicity experiment. Findings from the study indicate that when applied to drug-resistant clinical isolates and recombinant Mycobacterium smegmatis that expresses Rv1218c, two molecules – dodecanoic acid [DA] and palmitic acid [PA] – can lower the minimum inhibitory concentration [MIC] of Rifampicin by eight to a thousand time.43
Five small molecules – shikimic acid, rutin, myoinositol, methyl stearate, and ellagic acid – were discovered in other investigations 44,45 to work synergistically well with rifampicin to inhibit resistant M. tuberculosis isolates.46 A notable decrease in the minimum inhibitory concentration of rifampicin was observed, and both myoinositol and methyl stearate were shown to be entirely non-toxic to hematological and epithelial cells from different organs in ex vivo settings. Consequently, these substances may enhance the effectiveness of first-line anti-TB medications 4748 reports that Vanoxerine [GBR12909] acts as a disruptor of the membrane electric potential and can inhibit the efflux and growth of mycobacteria. It was observed that vanoxerine exhibits no detectable resistance, indicating that there is no specific protein target involved.
A number of lines of evidence point to the involvement of mycobacterial drug efflux pumps in the drug tolerance induced by macrophages.49 When tested for drug efflux, it was found that M. tuberculosis transfers the first-line antitubercular medicine rifampicin via a proton gradient-dependent mechanism. Moreover, they demonstrate that rifampicin efflux in M. tuberculosis is inhibited by verapamil, a recognized EPI that also blocks macrophage-induced rifampicin tolerance. Verapamil inhibits calcium channels, which also stops rifampicin efflux, just how it can make macrophages more tolerant. They observed that verapamil, with its human P-glycoprotein [PGP] – like inhibitory action, directly blocks the drug efflux pumps of M. tuberculosis.
In addition to verapamil, several efflux pump inhibitors were identified, including thioridazine, chlorpromazine, flupenthixol, and haloperidol. These compounds were tested in vitro and on human-infected macrophages against various drug-resistant strains of M. tuberculosis. Similar to verapamil, these drugs function as efflux inhibitors while also blocking ion channels. In vitro studies showed that each drug effectively inhibited active efflux and produced synergistic effects when used in combination with isoniazid and rifampicin. It was observed that antibiotic exposure, both in vitro and within macrophages, increased the expression of efflux genes in M. tuberculosis, regardless of the resistance class. Additionally, these drugs led to rapid and effective death of M. tuberculosis isolates, which was associated with a decrease in intracellular ATP levels. This suggests that the compounds reduced the intracellular mycobacterial load by activating lysosomal hydrolases and acidifying phagosomes.50
Unlike M. tuberculosis found in THP-1 macrophages, research has shown that M. tuberculosis can infect and thrive in bone marrow mesenchymal stem cells [BM-MSCs], which exhibit greater resistance to the anti-TB drugs isoniazid and rifampicin compared to the bacteria in THP-1 macrophages. The study also found that the ABCG2 efflux pumps in BM-MSCs were upregulated in response to rifampicin, potentially explaining the resistance of the bacteria within these cells to the antibiotics. Further investigations indicated that inhibiting ABCG2 efflux pumps with sorafenib, an efflux pump inhibitor [EPI], along with the administration of anti-TB drugs, increased the susceptibility of the bacteria, leading to enhanced death of M. tuberculosis in BM-MSCs. These findings demonstrated for the first time that M. tuberculosis residing in BM-MSCs significantly raises the minimum inhibitory concentration [MIC] of anti-TB medications.51
Conclusion
While EPIs may exhibit limited standalone efficacy, their ability to significantly enhance the potency and effectiveness of antibiotics and other therapeutics is increasingly evident. This synergistic potential positions EPIs as a compelling strategy in the fight against the growing threat of antibiotic resistance.
Research into EPIs as adjunct therapies underscores their promise as a viable approach to counter multi-drug-resistant (MDR) bacteria, a global challenge that has intensified the need for innovative solutions. The alarming rise of MDR pathogens has rendered many conventional antibiotics ineffective, necessitating novel interventions. EPIs stand out as a particularly promising avenue, offering a mechanism to restore the clinical utility of existing antibiotics and extend the lifespan of our pharmacological arsenal.
The strategic deployment of EPIs could not only prolong the effectiveness of current drugs but also pave the way for revitalizing antibiotics that have been sidelined due to resistance. A growing body of literature highlights the utility of EPIs as adjuvants in antibiotic therapy, demonstrating their capacity to circumvent resistance mechanisms. By inhibiting efflux pumps—key contributors to bacterial resistance—EPIs enhance antibiotic accumulation within bacterial cells, thereby amplifying therapeutic outcomes. Moreover, EPIs provide a foundation for developing innovative treatment paradigms that could fundamentally transform the management of bacterial infections.
In conclusion, EPIs represent a critical tool in the ongoing battle against antibiotic resistance. Their multifaceted properties, from enhancing drug efficacy to enabling novel therapeutic strategies, make them invaluable assets in addressing one of the most pressing public health challenges of our time. Continued research and clinical exploration of EPIs will be essential to fully harness their potential and reshape the future of antimicrobial therapy.
Acknowledgement
The authors would like to acknowledge the contributions of researchers worldwide whose studies on efflux pumps, efflux pump inhibitors, multi-drug resistance, and Mycobacterium tuberculosis have formed the foundation of this review.
Funding Sources
The author(s) received no financial support for the research, authorship, and/or publication of this article.
Conflict of Interest
The authors do not have any conflict of interest.
Data Availability Statement
This statement does not apply to this article.
Ethics Statement
This study did not involve human participants, and therefore, informed consent was not required.
Clinical Trial Registration
This research does not involve any clinical trials.
Permission to reproduce material from other sources
Not Applicable
Author Contributions
- Benjamin Isaac Thomson – Conceptualization, Writing, Editing – Original Draft.
- Shainaba A Saadhali – Analysis, Review & Editing.
References
- Gajic I, Jovicevic M, Popadic V, et al. The emergence of multi-drug-resistant bacteria causing healthcare-associated infections in COVID-19 patients: a retrospective multi-centre study. Journal of Hospital Infection. 2023;137:1-7. doi:10.1016/j.jhin.2023.04.013
CrossRef - Gaurav A, Bakht P, Saini M, Pandey S, Pathania R. Role of bacterial efflux pumps in antibiotic resistance, virulence, and strategies to discover novel efflux pump inhibitors. Microbiology (Reading). 2023;169(5). doi:10.1099/MIC.0.001333
CrossRef - Compagne N, Vieira Da Cruz A, Müller RT, Hartkoorn RC, Flipo M, Pos KM. Update on the Discovery of Efflux Pump Inhibitors against Critical Priority Gram-Negative Bacteria. Antibiotics (Basel). 2023;12(1). doi:10.3390/ANTIBIOTICS12010180
CrossRef - Hazra S, Hazarika R, Patra S. Multitargeting: An Alternative Approach to Tackle Multidrug Resistance in Tuberculosis. Curr Drug Targets. 2023;24(9):751-775. doi:10.2174/1389450124666230505145335
CrossRef - Huang L, Wu C, Gao H, et al. Bacterial Multidrug Efflux Pumps at the Frontline of Antimicrobial Resistance: An Overview. Antibiotics. 2022;11(4). doi:10.3390/ANTIBIOTICS11040520
CrossRef - Piddock LJV. Multidrug-resistance efflux pumps – Not just for resistance. Nat Rev Microbiol. 2006;4(8):629-636. doi:10.1038/NRMICRO1464.
CrossRef - Mohanty H, Pachpute S, Yadav RP. Mechanism of drug resistance in bacteria: efflux pump modulation for designing of new antibiotic enhancers. Folia Microbiol (Praha). 2021;66(5):727-739. doi:10.1007/S12223-021-00910-Z
CrossRef - AlMatar M, Albarri O, Makky EA, Köksal F. Efflux pump inhibitors: new updates. Pharmacological Reports. 2021;73(1). doi:10.1007/S43440-020-00160-9
CrossRef - Blair JMA, Webber MA, Baylay AJ, Ogbolu DO, Piddock LJV. Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol. 2015;13(1):42-51. doi:10.1038/nrmicro3380
CrossRef - Hernando-Amado S, Blanco P, Alcalde-Rico M, et al. Multidrug efflux pumps as main players in intrinsic and acquired resistance to antimicrobials. Drug Resistance Updates. 2016;28:13-27. doi:10.1016/J.DRUP.2016.06.007
CrossRef - Efflux Pump: Mechanism, Types, Role | Vaia. Accessed October 5, 2023. https://www.hellovaia.com/ explanations/biology/communicable-diseases/efflux-pump/
- Nishino K, Yamasaki S, Nakashima R, Zwama M, Hayashi-Nishino M. Function and Inhibitory Mechanisms of Multidrug Efflux Pumps. Front Microbiol. 2021;12. doi:10.3389/FMICB.2021.737288
CrossRef - Compagne N, Vieira Da Cruz A, Müller RT, Hartkoorn RC, Flipo M, Pos KM. Update on the Discovery of Efflux Pump Inhibitors against Critical Priority Gram-Negative Bacteria. Antibiotics (Basel). 2023;12(1). doi:10.3390/ANTIBIOTICS12010180
CrossRef - Ika Irianti M, Vincken JP, van Dinteren S, ter Beest E, Pos KM, Araya-Cloutier C. Prenylated isoflavonoids from Fabaceae against the NorA efflux pump in Staphylococcus aureus. Sci Rep. 2023;13(1). doi:10.1038/S41598-023-48992-8
CrossRef - Neuberger A, Du D, Luisi BF. Structure and mechanism of bacterial tripartite efflux pumps. Res Microbiol. 2018;169(7-8):401-413. doi:10.1016/J.RESMIC.2018.05.003
CrossRef - Dadashova B, Buehler R, Cherry C, Ye X. Equitable active transport. Transp Res D Transp Environ. 2023;119:103737. doi:10.1016/J.TRD.2023.103737
CrossRef - Luengo A, Li Z, Gui DY, et al. Increased demand for NAD+ relative to ATP drives aerobic glycolysis. Mol Cell. 2021;81(4):691-707.e6. doi:10.1016/J.MOLCEL.2020.12.012
CrossRef - Sharma A, Gupta VK, Pathania R. Efflux pump inhibitors for bacterial pathogens: From bench to bedside. Indian J Med Res. 2019;149(2):129. doi:10.4103/IJMR.IJMR_2079_17
CrossRef - Kumar S, Lekshmi M, Parvathi A, Ojha M, Wenzel N, Varela MF. Functional and structural roles of the major facilitator superfamily bacterial multidrug efflux pumps. Microorganisms. 2020;8(2). doi:10.3390/MICROORGANISMS8020266
CrossRef - Raj DS, Kesavan DK, Muthusamy N, Umamaheswari S. Efflux pumps potential drug targets to circumvent drug Resistance – Multi drug efflux pumps of Helicobacter pylori. Mater Today Proc. 2021;45:2976-2981. doi:10.1016/j.matpr.2020.11.955
CrossRef - Orelle C, Jault JM. Structures and Transport Mechanisms of the ABC Efflux Pumps. Efflux-Mediated Antimicrobial Resistance in Bacteria. Published online 2016:73-98. doi:10.1007/978-3-319-39658-3_4
CrossRef - Dhar J, Thai ALP, Ghoshal A, Giomi L, Sengupta A. Self-regulation of phenotypic noise synchronizes emergent organization and active transport in confluent microbial environments. Nature Physics 2022 18:8. 2022;18(8):945-951. doi:10.1038/s41567-022-01641-9
CrossRef - Nishino K, Yamasaki S, Nakashima R, Zwama M, Hayashi-Nishino M. Function and Inhibitory Mechanisms of Multidrug Efflux Pumps. Front Microbiol. 2021;12:737288. doi:10.3389/FMICB.2021. 737288/BIBTEX
CrossRef - Maloney FP, Kuklewicz J, Corey RA, et al. Structure, substrate recognition and initiation of hyaluronan synthase. Nature. 2022;604(7904):195-201. doi:10.1038/S41586-022-04534-2
CrossRef - Nies DH. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol Rev. 2003;27(2-3):313-339. doi:10.1016/S0168-6445(03)00048-2
CrossRef - Daury L, Orange F, Taveau JC, et al. Tripartite assembly of RND multidrug efflux pumps. Nat Commun. 2016;7. doi:10.1038/NCOMMS10731
CrossRef - Nikaido H. RND transporters in the living world. Res Microbiol. 2018;169(7-8):363-371. doi:10.1016/J.RESMIC.2018.03.001
CrossRef - Kuroda T, Tsuchiya T. Multidrug efflux transporters in the MATE family. Biochim Biophys Acta. 2009;1794(5):763-768. doi:10.1016/J.BBAPAP.2008.11.012
CrossRef - Lamut A, Peterlin Mašič L, Kikelj D, Tomašič T. Efflux pump inhibitors of clinically relevant multidrug resistant bacteria. Med Res Rev. 2019;39(6):2460-2504. doi:10.1002/MED.21591
CrossRef - Shriram V, Khare T, Bhagwat R, Shukla R, Kumar V. Inhibiting bacterial drug efflux pumps via phyto-therapeutics to combat threatening antimicrobial resistance. Front Microbiol. 2018;9(DEC). doi:10.3389/FMICB.2018.02990
CrossRef - Lomovskaya O, Warren MS, Lee A, et al. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: Novel agents for combination therapy. Antimicrob Agents Chemother. 2001;45(1):105-116. doi:10.1128/AAC.45.1.105-116.2001
CrossRef - Chevalier J, Atifi S, Eyraud A, Mahamoud A, Barbe J, Pagès JM. New pyridoquinoline derivatives as potential inhibitors of the fluoroquinolone efflux pump in resistant enterobacter aerogenes strains. J Med Chem. 2001;44(23):4023-4026. doi:10.1021/JM010911Z
CrossRef - Abdel-Karim SAAM, El-Ganiny AMA, El-Sayed MA, Abbas HAA. Promising FDA-approved drugs with efflux pump inhibitory activities against clinical isolates of Staphylococcus aureus. PLoS One. 2022;17(7 July). doi:10.1371/JOURNAL.PONE.0272417
CrossRef - Seukep AJ, Mbuntcha HG, Kuete V, Chu Y, Fan E, … What Approaches to Thwart Bacterial Efflux Pumps-Mediated Resistance? Antibiotics. Published online 2022. https://www.mdpi.com/2079-6382/11/10/1287
CrossRef - Seukep AJ, Fokoua-Maxime CD, Mbuntcha HG, et al. Bacterial Drug Efflux Pump Inhibitors from Plants. Antimicrobial Resistance: Underlying Mechanisms and Therapeutic Approaches. Published online January 1, 2022:487-532. doi:10.1007/978-981-16-3120-7_16
CrossRef - Dias KJSDO, Miranda GM, Bessa JR, et al. Terpenes as bacterial efflux pump inhibitors: A systematic review. Front Pharmacol. 2022;13. doi:10.3389/FPHAR.2022.953982
CrossRef - Blair JMA, Richmond GE, Piddock LJV. Multidrug efflux pumps in Gram-negative bacteria and their role in antibiotic resistance. Future Microbiol. 2014;9(10):1165-1177. doi:10.2217/FMB.14.66
CrossRef - Qaderi MM, Martel AB, Strugnell CA. Environmental Factors Regulate Plant Secondary Metabolites. Plants (Basel). 2023;12(3). doi:10.3390/PLANTS12030447
CrossRef - Noor F, Qamar MTU, Ashfaq UA, Albutti A, Alwashmi ASS, Aljasir MA. Network Pharmacology Approach for Medicinal Plants: Review and Assessment. Pharmaceuticals (Basel). 2022;15(5). doi:10.3390/PH15050572
CrossRef - Seukep AJ, Kuete V, Nahar L, Sarker SD, Guo M. Plant-derived secondary metabolites as the main source of efflux pump inhibitors and methods for identification. J Pharm Anal. 2020;10(4):277-290. doi:10.1016/j.jpha.2019.11.002
CrossRef - Sau S, Kalia NP. Perspectives on Anti-Tuberculosis Drug Discovery. Drugs and a Methodological Compendium. Published online 2023:357-375. doi:10.1007/978-981-19-7952-1_13
CrossRef - Laws M, Jin P, Rahman KM. Efflux pumps in Mycobacterium tuberculosis and their inhibition to tackle antimicrobial resistance. Trends Microbiol. 2022;30(1):57-68. doi:10.1016/J.TIM.2021.05.001
CrossRef - Nirmal CR, Rajadas SE, Balasubramanian M, et al. Dodecanoic acid & palmitic acid disarms rifampicin resistance by putatively targeting mycobacterial efflux pump Rv1218c. Indian Journal of Medical Research. 2023;157(23):192-203. doi:10.4103/IJMR.IJMR_1610_22
CrossRef - Rajput VS, Runthala A, Khan IA. Shikimate Kinase Inhibitors: An Update on Promising Strategy against Mycobacterium tuberculosis . Curr Drug Targets. 2023;24(5):388-405. doi:10.2174/ 1389450124666230208102645
CrossRef - Kawamoto S, Hori C, Taniguchi H, Okubo S, Aoki S. Identification of novel antimicrobial compounds targeting Mycobacterium tuberculosis shikimate kinase using in silico hierarchical structure-based drug screening. Tuberculosis. 2023;141. doi:10.1016/J.TUBE.2023.102362
CrossRef - Mehra R, Rajput VS, Gupta M, et al. Benzothiazole Derivative as a Novel Mycobacterium tuberculosis Shikimate Kinase Inhibitor: Identification and Elucidation of Its Allosteric Mode of Inhibition. J Chem Inf Model. 2016;56(5):930-940. doi:10.1021/ACS.JCIM.6B00056/SUPPL_FILE/CI6B00056_SI_001.PDF
CrossRef - Nirmal CR, Rajadas SE, Balasubramanian M, et al. Myoinositol and methyl stearate increases rifampicin susceptibility among drug-resistant Mycobacterium tuberculosis expressing Rv1819c. Chem Biol Drug Des. 2023;101(4):883-895. doi:10.1111/CBDD.14197
CrossRef - Kingdon ADH, Meosa-John AR, Batt SM, Besra GS. Vanoxerine kills mycobacteria through membrane depolarization and efflux inhibition. Front Microbiol. 2023;14:1112491. doi:10.3389/ fmicb.2023. 1112491
CrossRef - Lake MA, Adams KN, Nie F, et al. The human proton pump inhibitors inhibit Mycobacterium tuberculosis rifampicin efflux and macrophage-induced rifampicin tolerance. Proc Natl Acad Sci U S A. 2023;120(7). doi:10.1073/PNAS.2215512120
CrossRef - Machado D, Pires D, Perdigão J, et al. Ion channel blockers as antimicrobial agents, efflux inhibitors, and enhancers of macrophage killing activity against drug resistant mycobacterium tuberculosis. PLoS One. 2016;11(2). doi:10.1371/journal.pone.0149326
CrossRef - Kaur S, Angrish N, Gupta K, Tyagi AK, Khare G. Inhibition of ABCG2 efflux pumps renders the Mycobacterium tuberculosis hiding in mesenchymal stem cells responsive to antibiotic treatment. Infect Genet Evol. 2021;87. doi:10.1016/J.MEEGID.2020.104662
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





