Manuscript accepted on : 25-09-2025
Published online on: 30-09-2025
Plagiarism Check: Yes
Reviewed by: Dr. Per A, Löthman
Second Review by: Dr. Gopal Samy and Dr. Rajesh Damodharan
and Dr. Rajesh Damodharan
Final Approval by: Dr. Eugene A. Silow
CRISPR/CAS9-Mediated Gene Editing in Human Gametes: A Review
Esha Kumari1 , Neha Banu1
, Neha Banu1 , Kathrina Marbaniang2
, Kathrina Marbaniang2 , Faridha Jane R.M. Momin3
, Faridha Jane R.M. Momin3 and Barry Cooper Hynniewta4*
and Barry Cooper Hynniewta4*
1Department of Clinical Embryology, Yenepoya (deemed to be university), Karnataka, India
2Department of Medicine, Shillong Medical College, Meghalaya, India
3Department of Oncology, Shillong Civil Hospital, Meghalaya, India
4Department of Clinical Embryology, MOMSOON Fertility and IVF Centre, Karnataka, India
Corresponding Author E-mail:barrycooperhynniewta@gmail.com
ABSTRACT: Gene editing exploits endogenous DNA repair pathways to introduce precise modifications into the genome. The CRISPR system was first identified in Escherichia coli (1987) and, based on Cas protein architecture, is divided into Class I (types I, III, IV) and Class II (types II, V, VI). The widely adopted CRISPR/Cas9 system comprises a guide RNA (gRNA) and the Cas9 endonuclease, which orchestrate genome editing through a tripartite mechanism: target sequence recognition via gRNA binding to a complementary DNA site adjacent to a protospacer adjacent motif (PAM), double-stranded DNA cleavage by two nuclease domains (RuvC cleaving the non‑target strand and HNH the target strand), and repair of the resulting break by cellular pathways. This approach has been applied to in vitro fertilization-derived embryos and meiotically developing oocytes to disrupt or correct genes, offering potential for eliminating heritable diseases. However, concerns remain regarding off-target effects that introduces unwanted genetic mutations, necessitating improved specificity and ethical scrutiny.
KEYWORDS: CRISPR/Cas9; DNA; Gametes; Gene editing; Infertility
| Copy the following to cite this article: Kumari E, Banu N, Marbaniang K, Momin F. J. R. M, Hynniewta B. C. CRISPR/CAS9-Mediated Gene Editing in Human Gametes: A Review. Biotech Res Asia 2025;22(3). |
| Copy the following to cite this URL: Kumari E, Banu N, Marbaniang K, Momin F. J. R. M, Hynniewta B. C. CRISPR/CAS9-Mediated Gene Editing in Human Gametes: A Review. Biotech Res Asia 2025;22(3). Available from: https://bit.ly/48avFIP |
Introduction
Gene editing employs endogenous DNA repair mechanisms to introduce precise, targeted modifications into the human genome, significantly advancing genetic research. Gene editing is a process that make use of the biochemical processes that naturally repair DNA damage. Various nuclease mechanisms are able to introduce DNA breaks at specified sites in the genome. The Cas9–sgRNA gene editing platform, which is based on the bacterial adaptive CRISPR immune system, has gained attention in gene editing recently and is now a standard procedure in many research laboratories across the globe due to the ease of use.1 Recombinant DNA technology is a set of techniques used to recombine (join) DNA Segments of two or more distinct DNA molecules which are put together to produce a recombinant DNA molecule. A recombinant DNA molecule is able to enter a cell and replicate there under specific conditions, either by itself or by following chromosome integration.2 Any species provide DNA sequences that are used to generate recombinant DNA molecules. In some cases, bacterial and plant DNA are put together, or fungus and human DNA are mixed. The chemically-mediated creation of DNA also produces DNA sequences that are not found in nature and incorporate them into recombinant molecules. It is possible to create and introduce any DNA sequence into a wide range of living organisms through the use of synthetic DNA and recombinant DNA technology.3
Infertility
A reproductive disorder known as infertility that affects both sexes and is marked by infertility following a period of regular, unprotected sexual activity lasting twelve months or longer.4 It is classified into two types: primary infertility, which occurs when there has never been a successful pregnancy, and secondary infertility, which occurs when there has been at least one successful pregnancy but still no baby.
Male Infertility
Male infertility is typically characterized by issues with sperm ejection, low sperm count, or poor morphology and motility.5 Spermatogenesis comprises a sequence of cellular processes, such as mitosis-induced self-renewal of spermatogonial stem cells and spermatogonia, spermatocyte transformation and differentiation, meiosis I/II-induced haploid spermatid generation, and spermiogenesis-induced final morphological maturation of spermatids to become spermatozoa. Thus, these processes entail a variety of cellular activities in the testis and are intricately regulated by signalling and hormonal axes.6-10
Female Infertility
Infertility in the female reproductive system originate from problems in the ovaries, uterus, fallopian tubes, and endocrine system, among other factors5. In the female embryo, a number of intraovarian and extraovarian components regulate the highly complex process of creating gametes, leading to a progeny organism.11 The process through which the oocyte, is formed is known as oogenesis. There are many interactions between the developing oocyte and the granulosa and cumulus cells that surround it in this multi-step process. When oogonia are formed from primordial germ cells (PGC), about the 12th week of a woman’s pregnancy, oogenesis starts in the foetal ovaries as soon as the embryo’s development expands.12-15
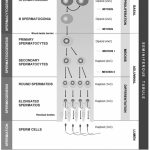 |
Figure 1: Flow chart representing stages of spermatogenesis, germ cell characteristics in each stage and compartments of seminiferous tubules.16 |
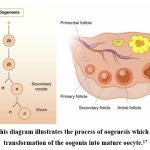 |
Figure 2: This diagram illustrates the process of oogenesis which begins with transformation of the oogonia into mature oocyte.17 |
Evolution and Milestones of Crispr Technology
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) were initially identified in the genomic DNA of Escherichia coli by Ishino et al. (1987) at Osaka University, Japan.18 Subsequently, in 1995, Francisco Mojica, a Spanish microbiologist, significantly advanced the understanding of CRISPR loci by identifying analogous sequences in the archaeal genome of Haloferax mediterranei, thereby suggesting a conserved function across prokaryotic domains.19 The first experimental evidence elucidating the functional mechanism of the CRISPR-Cas system emerged in 2007 through studies conducted on Streptococcus thermophilus, a bacterium used in yogurt fermentation. This research was led by Rodolphe Barrangou and Philippe Horvath under the auspices of the Danish biotechnology firm Danisco.20 Since the 1980s, Danisco had developed an extensive repository of bacterial strains, enabling detailed analyses of bacteriophage-host interactions. These investigations revealed the adaptive nature of the CRISPR system, wherein the incorporation of novel spacer sequences into the CRISPR locus correlated with acquired immunity against corresponding bacteriophages. Building on these findings, the researchers pioneered a CRISPR-based strategy for bacterial immunization, culminating in one of the earliest patents in this domain, filed in 2005.21,22 The CRISPR-Cas9 system relies on a small RNA molecule that is processed and transcribed from the CRISPR locus. This molecule directs Cas proteins to external nucleic acid sequences that contain identical genetic information. A study group headed by John van der Oost at Wageningen University in the Netherlands was the first to identify these RNA components, which are called CRISPR RNAs (crRNAs).23 Essential for in vitro reconstituting of the CRISPR-Cas9 system, an extra short RNA molecule involved in crRNA maturation was found in 2011 by Emmanuelle Charpentier’s lab. The activation of Cas9 nuclease activity was demonstrated to require this RNA, which is known as trans-activating CRISPR RNA (tracrRNA). A significant step towards the programmable and practical use of the CRISPR-Cas9 system for genome editing was the conceptual advancement that crRNA and tracrRNA are created into a single chimeric RNA, called single-guide RNA (sgRNA).24,25
 |
Figure 3: The historical chronology of the CRISPR-Cas9 system’s component findings.26 |
Components of CRISPR
CRISPR/Cas systems are broadly classified into two major classes based on the structural organization and functional characteristics of their associated Cas proteins: Class I, comprising types I, III, and IV, and Class II, comprising types II, V, and VI.27 Class I systems utilize multi-subunit protein complexes to mediate interference, whereas Class II systems rely on a single, multifunctional Cas protein to perform the same function. Two fundamental elements are required for the CRISPR/Cas9 system to function: the Cas9 endonuclease and the guide RNA (gRNA). Genome editing initially made use of the Cas9 protein, which originated from Streptococcus pyogenes (SpCas9). The capacity to produce specific double-stranded DNA breaks at specific places has earned this big, multidomain protein, which contains 1,368 amino acids, the nickname “molecular scissor”.28 The two primary structural domains of Cas9 are the recognition (REC) and nuclease (NUC) domains. The REC lobe, which is made up of the REC1 and REC2 domains, helps with target recognition by binding the guide RNA. A number of domains are located in the NUC lobe. One of these is the RuvC nuclease domain, which cleaves the non-target DNA strand. The other is the protospacer adjacent motif (PAM)-interacting domain, which binds to the target DNA’s PAM sequence and guarantees sequence specificity.29 An 18–20 nucleotide sequence complementary to the target DNA is found in the CRISPR RNA (crRNA) component of the guide RNA, while a scaffold of stem-loop structures required for Cas9 binding and activation is formed by the trans-activating CRISPR RNA (tracrRNA).28 These RNAs work in tandem to enable site-specific DNA cleavage by directing the Cas9 protein to particular genomic locations.
CRISPR–CAS System Mechanism
The three main steps of the CRISPR/Cas9 system for editing genomes are identifying targets, cleaving DNA, and repairing damaged DNA.30 Synthetic single-guide RNAs (sgRNAs) contain CRISPR RNA (crRNA) components that base-pair with complementary sequences in the target gene, directing the Cas9 endonuclease to the target DNA. When sgRNA is not present, Cas9 stays in an inactive state. Cas9 will insert a double-strand break (DSB) at a position three nucleotides before the protospacer adjacent motif (PAM)31 when base pairing is successful. Although the precise sequence and length of the PAM differ among bacterial species, it is usually a brief, conserved DNA sequence located just downstream of the target site and usually ranging from 2 to 5 base pairs in length. The most popular version used for genome editing, SpCas9, which is derived from Streptococcus pyogenes, identifies the PAM sequence 5ʹ-NGG-3ʹ. Despite our limited understanding of the exact molecular mechanism, Cas9 is able to unwind local DNA strands and generate an RNA-DNA heteroduplex by recognising the PAM site. The catalytic domains of Cas9 are activated by this conformational shift. The DNA strand that is complementary to the sgRNA is cleaved by the HNH domain, and the DSBs that are caused by the RuvC domain are mainly blunt-ended. The endogenous DNA repair pathways of the host cell then fix these breaks, finishing the genome editing process.28,32
 |
Figure 4: This represents successive steps of CRISPR/Cas9 system which includes target identification and modification of gene.33 |
Some DNA strands experience double-strand breaks (DSBs). The crRNA directs Cas9. TracrRNA stabilizes a structure before Cas9 enabling to break the target DNA. sgRNA is responsible for target gene recognition [sgRNA (teal)].34 These domains (RuvC, HNH) then interact with the Cas proteins to effectively modify the genomes of different animals by introducing DSBs in the DNA at specific locations. The HNH (histidine–asparagine–histidine) nuclease domain of Cas9 cleaves the DNA strand complementary to the guide RNA, whereas the RuvC-like (resolvase C-like) nuclease domain cleaves the non-complementary strand.35 In both type I and type II CRISPR/Cas systems, the target of interference is foreign DNA containing a protospacer adjacent motif (PAM), a short, conserved sequence essential for target recognition and cleavage. The Cas9 protein breaks the target DNA on both strands using its RuvC and (HNH) domains. For Cas9 to break DNA, it primarily requires the PAM sequence. A 20-base stretch offers selectivity for binding in sgRNA. There are two methods for repairing DNA DSBs: HDR, which requires the presence of a template which results in NHEJ, a loose but permanent knockout of a gene, or knock-in or gene replacement.36 In type I and type II CRISPR/Cas systems, only foreign DNA sequences containing a protospacer adjacent motif (PAM)—a short, conserved nucleotide sequence—are specifically recognized and targeted for interference.37
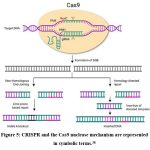 |
Figure 5: CRISPR and the Cas9 nuclease mechanism are represented in symbolic terms.38 |
The basic mechanism by which all living things sustain their generations is reproduction. Recently, a novel, adaptable genome editing technique called CRISPR/Cas9 was developed to fix genetic abnormalities that cause a number of diseases, expanding its ability to enhance reproductive health.39 Using CRISPR and ARTs together has made it easier to modify the genomes of embryos created using IVF and other similar procedures. CRISPR/Cas9 is especially useful when it comes to IVF. IVF-produced embryos have the potential to have particular genes disrupted or edited by CRISPR, which could improve particular features or prevent hereditary diseases.40 In general, CRISPR technology has the capacity to improve IVF results and encouraging the development of new uses for the reproductive systems of both men and women.41
Application of CRISPR in Reproductive Biology
SPERM
Genome editing technologies have the ability to revolutionize spermatogenesis research and shed light on the molecular mechanisms behind male infertility disorders. This was demonstrated by the successful application of CRISPR/Cas9-mediated gene editing in mouse spermatogonial stem cells (SSCs).42 An effective and easily accessible in vivo method for studying gene function during spermatogenesis is the CRISPR/Cas9-based spermatogenic cell-specific knockdown system.43 A proof of concept was carried out in rat SSCs using targeted gene editing at the Epsti1 locus. This locus is involved in epithelial-stromal interactions, and the results showed that the genome could be effectively modified. Epsti1 mutations in humans have been linked to changes in sperm function, which is worth noting.44 With SSCs readily available, a potential platform for homology-directed repair (HDR) mediated by CRISPR/Cas9 to address harmful mutations has emerged. This method has been proven effective in a model of cataract in mice by restoring normal gene function after an ex vivo correction of a disease-causing mutation in SSCs using CRISPR-Cas9 and HDR.45-47 These results raise the possibility that recovering spermatogenesis in individuals with genetically-caused non-obstructive azoospermia (NOA) could be possible through targeted gene editing in SSCs.48
OOCYTE
Generating genetically modified female germ cells for use in assisted reproductive technologies (ARTs) has been made possible by the CRISPR/Cas9 system, which has shown great promise in genetically modifying developing oocytes. These changes could be useful in preventing off-target mutations from being passed down across generations. Oocytes are a good candidate for germline modification because they are easily accessible. To test the efficacy and accuracy of CRISPR/Cas9-mediated gene editing, oocytes must first reach the germinal vesicle (GV) stage of in vitro maturation, and then the following meiotic development is required.39
Numerous animal models, including pigs and mice, have demonstrated the successful use of CRISPR/Cas9 to mammalian oocytes and embryos. It is possible to treat hereditary diseases by directly modifying the genome at these early phases of development. To further our understanding of the molecular pathways essential for embryogenesis, genome editing techniques in oocytes and embryos enable in-depth mechanistic research of early developmental processes.49
The dCas9-DNMT and dCas9-TET complexes are epigenome editing tools that have been used to alter DNA methylation patterns in embryos and oocytes from mammals. To fix aberrant methylation linked to familial Angelman syndrome, for instance, microinjected dCas9-TET-based systems have been used in mice oocytes.50 Furthermore, a CRISPR-based approach integrating sgRNA and dCas9-DNMT3a has been used to successfully edit seven separate genomic imprinting loci in single unfertilised oocytes, leading to the production of genetically changed children after fertilisation.51 In light of these developments, studies investigating oocyte methylation have gained popularity as a possible strategy for the treatment of non-genetic maternal hereditary disorders.52
Researchers have shown that injecting Cas9 cRNA into metaphase II (mII) oocytes before sperm and guide RNA (gRNA) has the capacity to increase editing efficiency, prolong Cas9 expression, and improve the results of genome modification. Edited embryos and healthy offspring could be produced using this co-injection technique.53 To further emphasise the significance of timing genome editing with DNA synthesis and particular cell cycle stages for optimal efficiency, it was found that inserting CRISPR/Cas9 components into M-phase oocytes successfully removed mosaicism in cleaving embryos.54
Even in mature oocytes with condensed chromatin, CRISPR/Cas9 was able to elicit high-efficiency alterations, according to the experimental findings. Gene editing is not the best approach for GV-stage oocytes, but they are made more mutable by blocking nuclear export and raising nuclear Cas9 levels. There were no negative impacts on meiotic progression or early embryonic development observed in pig models when CRISPR components were microinjected into immature oocytes.55-59 Genome editing in oocytes going through meiosis confirmed their eligibility as targets for CRISPR/Cas9-based therapies, as it led to a noticeably increased mutation efficiency.60,61
EMBRYO
To correct pathogenic mutations in the germline, an optimized CRISPR/Cas9-based strategy has been developed that leverages the endogenous DNA repair mechanisms active in early embryos. This approach enabled the precise, efficient, and accurate correction of a heterozygous mutation in the MYBPC3 gene—implicated in hypertrophic cardiomyopathy (HCM)—in human preimplantation embryos. This was achieved by co-injecting sperm, Cas9 protein, guide RNA (gRNA), and single-stranded oligodeoxynucleotides (ssODNs) into metaphase II (MII) oocytes, without inducing significant off-target effects or large deletions.62-64
When it comes to in vitro fertilization and preimplantation genetic diagnosis, the CRISPR/Cas9 method could be used to increase the quantity of embryos available for transfer.65 The use of CRISPR on human embryos has the potential for removing all genetic defects from the genome.66.
Advantages of CRISPR
Mutations are induced on several locations at once with the use of CRISPR/Cas9. Previously, this couldn’t be accomplished in a single round using traditional methods.67,68 It was necessary to create several mutant mouse lines using these traditional techniques, which necessitated repeated crossbreeding to produce mice with various mutations for study. As established by Wang et al., (2013) who simultaneously targeted the Tet1 and Tet2 genes, CRISPR/Cas9 may target multiple locations.67 Because CRISPR/Cas9 has the potential to create homozygous mice in the founding generation, analysis completed much more quickly and researchers are capable to generate more data than ever before by expanding their list of target genes.69
In the field of reproductive biology, the development of CRISPR/Cas9 technology is very encouraging. It is possible to thoroughly study the functional roles of potential genes implicated in spermatogenesis by using CRISPR/Cas9-mediated transcriptional suppression. This method allows for the assessment of gene-specific roles in sperm maturation. Furthermore, by fusing CRISPR/Cas9 systems with fluorescent tags, it is possible to establish the chromosomal localisation of genes specific to sperm. By making it easier to see and map spermatogenesis-related gene loci, these methods provide light on important regulatory mechanisms supporting male reproductive function.70
Limitations
One of the major challenges in applying CRISPR/Cas9 for gene therapy is the high frequency of off-target effects (OTEs), which have been reported at rates of ≥50% in some studies.71 Additional limitations of the CRISPR/Cas9 system include unintended on-target effects, suboptimal homology-directed repair (HDR) efficiency, and the persistent difficulty of precisely controlling genome edits.72–74 These issues raise concerns about the genomic integrity of edited embryos, as unanticipated mutations carry the risk of having deleterious consequences for progeny. As CRISPR technology continues to evolve, previously unrecognized editing errors continue to emerge, highlighting the need for comprehensive assessment and refinement prior to clinical application.75,76
Furthermore, embryos exhibit a distinct response to CRISPR-Cas9-induced DNA damage compared to somatic cells, a phenomenon not yet fully understood. Editing outcomes in embryos remain highly variable and unpredictable, frequently resulting in diverse forms of genomic damage. Notably, approximately 50% of edited embryonic cells display detectable abnormalities, indicating a high burden of unintended effects. These findings suggest that genome editing protocols optimized for somatic cells are not directly transferable to embryos. The induction of double-strand breaks (DSBs) by Cas9 in embryonic genomes occasionally leads to undetected or inaccurately repaired lesions due to incomplete knowledge of embryonic DNA repair pathways. Collectively, these challenges underscore the current unsuitability of CRISPR-Cas9 for clinical germline genome editing and emphasize the need for further investigation and ethical deliberation before its application in mammalian eggs and embryos.49,77
Bioethics
An important ethical consideration with CRISPR/Cas9 is the possibility of off-target consequences leading to alterations that were not intended. If such changes occur in germline cells, they could be passed on to future generations, potentially leading to unknown biological consequences. CRISPR/Cas9 safety needs to be improved by increasing its specificity and carefully identifying both desirable edits and off-target modifications. Another issue is mosaicism, in which certain cells are not altered, resulting in mutant cells that still cause illness and decreasing the efficacy of treatment.78 As mentioned earlier, the existing guidelines indicate that CRISPR technology should not be utilized for genetic editing in germ cells or embryos intended for implantation due to the uncertain and serious possible repercussions of off-target editing, the implications of modifying the intended target itself, the emergence of mosaicisms (the existence of two distinct cell lines derived from the same zygote), and the potential creation of new diseases along with the risk of impacting an entire generation of humans with these conditions.79
Conclusion
CRISPR/Cas9 technology holds transformative potential in reproductive biology, particularly in the genetic editing of human gametes. Because of its accuracy, effectiveness, and capacity to target certain DNA sequences, it is a useful tool for repairing genetic abnormalities at the gamete or zygote stage. Pre-fertilization gene editing has shown encouraging results in applications in spermatogonial stem cells (SSCs) and metaphase II oocytes, setting the stage for preventing monogenic hereditary diseases and enhancing embryo quality. Furthermore, improving embryo selection and combining CRISPR with methods such as preimplantation genetic diagnosis (PGD) are expected to greatly increase the success rates of assisted reproductive technologies (ART).
Although it offers many advantages, significant ethical and technical barriers still prevent CRISPR from being used clinically in reproductive medicine. There is considerable safety issues related to embryonic mosaicism, off-target effects, and unusual genomic rearrangements. Long-term follow-up research and strict regulatory frameworks are also necessary due to the possibility of irreversible germline changes. It is also necessary to carefully consider the ethical concerns of heredity, consent for future generations, and institutional concerns. Future research efforts should prioritize the optimization of target specificity, the minimization of associated risks, and the facilitation of transparent ethical discourse to guide the safe and responsible clinical implementation of CRISPR/Cas9 in reproductive health.
Acknowledgement
We would like to sincerely thank the Principal of Momsoon Academy for his continuous support and encouragement throughout the course of this research work.
Funding Sources
The authors received no financial support for the research, authorship, and/or publication of this article.
Conflict of Interest
The authors do not have any conflict of interest.
Data Availability Statement
This statement does not apply to this article.
Ethics Statement
This research did not involve human participants, animal subjects, or any material that requires ethical approval.
Informed Consent Statement
This study did not involve human participants, animal subjects, or any material that requires ethical approval.
Clinical Trial Registration
This research does not involve any clinical trials.
Permission to Reproduce Material from Other Sources
Not Applicable.
Authors Contribution
Esha Kumari: Conceptualization, Methodology, Writing- Original Draft.
Neha Banu: Data Collection, Analysis, Writing- Review and Editing.
Katrina Marbaniang: Analysis, Editing and Supervision.
Faridha Jane R.M. Momin: Analysis, Editing and Supervision.
Barry Cooper Hynniewta: Visualization, Supervision, Project Administration.
References
- Bak RO, Gomez-Ospina N, Porteus MH. Gene editing on center stage. Trends Genet. 2018 Aug;34(8):600–11. doi: 10.1016/j.tig.2018.05.004.
CrossRef - GNN- Genetics and Genomics Timeline”. www.genomenewsnetwork.org. Retrieved 16 February 2018.
- Rosano, GL.; Ceccarelli, EA. (2014-04-17). “Recombinant protein expression in Escherichia coli: advances and challenges”. Frontiers in Microbiology. doi:10.3389/fmicb. 2014.00172. ISSN 1664-302X. PMC 4029002 . PMID 24860555.
CrossRef - World Health Organization (WHO). International Classification of Diseases, 11th Revision (ICD-11). Geneva: WHO; 2018.
- World Health Organization. WHO fact sheet on infertility. Glob Reprod Health. 2021;6:e52. doi:10.1097/GRH.0000000000000052.
CrossRef - O’Donnell L, Robertson KM, Jones ME, Simpson ER. Estrogen and spermatogenesis. Endocr Rev. 2001 Jun;22(3):289-318. doi: 10.1210/edrv.22.3.0431. PMID: 11399746.
CrossRef - McLachlan RI, O’Donnell L, Meachem SJ, Stanton PG, de K, Pratis K, Robertson DM. Hormonal regulation of spermatogenesis in primates and man: insights for development of the male hormonal contraceptive. J Androl. 2002 Mar-Apr;23(2):149-62. PMID: 11868805.
CrossRef - Carreau S, Hess RA. Oestrogens and spermatogenesis. Philos Trans R Soc Lond B Biol Sci. 2010 May 27;365(1546):1517-35. doi: 10.1098/rstb.2009.0235. PMID: 20403867; PMCID: PMC2871919.
CrossRef - Sharpe RM. Regulation of spermatogenesis. In: The Physiology of Reproduction. Eds. Knobil, E., Neill, J.D. New York, Raven Press. 1994. pp. 1363–434.
- O’Donnell L, Meachem SJ, Stanton PG, McLach lan RI. Endocrine regulation of spermatogenesis. In: Neill JD, editor. Physiology of Reproduction. 3rd ed. Amsterdam: Elsevier; 2006. p. 1017–69.
CrossRef - Sánchez F, Smitz J. Molecular control of oogenesis. Biochim Biophys Acta. 2012 Dec;1822(12):1896-912. doi: 10.1016/j.bbadis.2012.05.013. Epub 2012 May 24. PMID: 22634430.
CrossRef - Virant-Klun I. Postnatal oogenesis in humans: a review of recent findings. Stem Cells Cloning. 2015 Mar 20;8:49-60. doi: 10.2147/SCCAA.S32650. PMID: 25848307; PMCID: PMC4376261.
CrossRef - Yatsenko SA, Rajkovic A. Genetics of human female infertility†. Biol Reprod. 2019 Sep 1;101(3):549-566. doi: 10.1093/biolre/ioz084. PMID: 31077289; PMCID: PMC8127036.
CrossRef - Machtinger R, Laurent LC, Baccarelli AA. Extracellular vesicles: roles in gamete maturation, fertilization and embryo implantation. Hum Reprod Update. 2016 Mar-Apr;22(2):182-93. doi: 10.1093/humupd/dmv055. Epub 2015 Dec 9. PMID: 26663221; PMCID: PMC4755440.
- MacLennan M, Crichton JH, Playfoot CJ, Adams IR. Oocyte development, meiosis and aneuploidy. Semin Cell Dev Biol. 2015 Sep;45:68-76. doi: 10.1016/j.semcdb.2015.10.005. Epub 2015 Oct 8. PMID: 26454098; PMCID: PMC4828587.
CrossRef - Salido GM, Rosado JA, editors. Apoptosis: involvement of oxidative stress and intracellular Ca²⁺ homeostasis. Dordrecht: Springer; 2009. doi:10.1007/978-1-4020-9873-4.
CrossRef - Yao X, Liu W, Xie Y, Xi M, Xiao L. Fertility loss: negative effects of environmental toxicants on oogenesis. Front Physiol. 2023; 14:1219045. doi:10.3389/fphys.2023.1219045.
CrossRef - Ishino, Y., Shinagawa, H., Makino, K., Amemura, M., and Nakatura, A. (1987) Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isoenzyme conversion in Escherichia coli, and identification of the gene product, Bacteriol., 169, 5429-5433, doi: 10.1128/ jb.169.12.5429-5433.1987.
CrossRef - Mojica, F. J. M., Juez, G., and Rodriguez‐Valera, F. (1993) Transcription at different salinities of Haloferax mediterranei sequences adjacent to partially modified PstI sites, Microbiol., 9, 613-621, doi: 10.1111/j.1365-2958.1993.tb01721.x.
CrossRef - Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. 2007 Mar 23;315(5819):1709-12. doi: 10.1126/science.1138140. PMID: 17379808.
CrossRef - Horvath, P., Barrangou, R., Fremaux, C., Boyaval, P., and Romero, D. (2007) Use of a Cas Gene in Combination with CRISPR Repeats for Modulating Resistance in a Cell. US Patent Application No. PCT/US2006/033167 (initially filed on 26.08.2005).
- Isaacson, W. (2021) The Code Breaker: Jennifer Doudna, Gene Editing, and the Future of the Human Race, Simon & Schuster (New York, USA).
- Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008 Aug 15;321(5891):960-4. doi: 10.1126/science.1159689. PMID: 18703739; PMCID: PMC5898235.
CrossRef - Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. 2011 Mar 31;471(7340):602-7. doi: 10.1038/nature09886. PMID: 21455174; PMCID: PMC3070239.
CrossRef - Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doud na, J. A., et al. (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity, Science, 337, 816-821, doi: 10.1126/science.1225829.
CrossRef - Gostimskaya I. CRISPR-Cas9: A History of Its Discovery and Ethical Considerations of Its Use in Genome Editing. Biochemistry (Mosc). 2022 Aug;87(8):777-788. doi: 10.1134/S0006297922080090. PMID: 36171658; PMCID: PMC9377665.
CrossRef - Liu Z, Dong H, Cui Y, Cong L, Zhang D. Application of different types of CRISPR/Cas-based systems in bacteria. Microb Cell Fact. 2020;19(1):1–14. doi:10.1186/s12934-020-01431-z
CrossRef - Mei Y, Wang Y, Chen H, Sun ZS, Da JX. Recent progress in CRISPR/Cas9 technology. J Genet Genomics. 2016;43(2):63–75. doi:10.1016/j.jgg.2016.01.001
CrossRef - Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. 2014 Feb 27;156(5):935-49. doi: 10.1016/j.cell.2014.02.001. Epub 2014 Feb 13. PMID: 24529477; PMCID: PMC4139937.
CrossRef - Shao M, Xu TR, Chen CS. The big bang of genome editing technology: development and application of the CRISPR/Cas9 system in disease animal models. Dongwuxue Yanjiu. 2016 Jul 18;37(4):191-204. doi: 10.13918/j.issn.2095-8137.2016.4.191. PMID: 27469250; PMCID: PMC4980067.
- Ceasar SA, Rajan V, Prykhozhij SV, Berman JN, Ignacimuthu S. Insert, remove or replace: a highly advanced genome editing system using CRISPR/Cas9. Biochim Biophys Acta Mol Cell. 2016;1863 (9):2333–2344. doi:10.1016/j.bbamcr.2016.06.009
CrossRef - . Jiang F, Doudna JA. CRISPR-Cas9 structures and mechanisms. Annu Rev Biophys. 2017;46(1):505–529. doi:10.1146/annurev-biophys -062215-010822
CrossRef - Aljabali AAA, El-Tanani M, Tambuwala MM. Principles of CRISPR-Cas9 technology: Advancements in genome editing and emerging trends in drug delivery. J Drug Deliv Sci Technol. 2024; 92:105338. doi: 10.1016/j.jddst.2023.105338.
CrossRef - Manghwar H, Lindsey K, Zhang X, Jin S. CRISPR/Cas System: Recent Advances and Future Prospects for Genome Editing. Trends Plant Sci. 2019 Dec;24(12):1102-1125. doi: 10.1016/j.tplants.2019.09.006. Epub 2019 Nov 11. PMID: 31727474.
CrossRef - Hu JH, Miller SM, Geurts MH, Tang W, Chen L, Sun N, Zeina CM, Gao X, Rees HA, Lin Z, Liu DR. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature. 2018 Apr 5;556(7699):57-63. doi: 10.1038/nature26155. Epub 2018 Feb 28. PMID: 29512652; PMCID: PMC5951633.
CrossRef - Kundar R, Gokarn K. CRISPR-Cas System: A Tool to Eliminate Drug-Resistant Gram-Negative Bacteria. Pharmaceuticals. 2022; 15(12):1498. https://doi.org/10.3390/ph15121498
CrossRef - Das A, Goswami HN, Whyms CT, Sridhara S and Li H, “Structural Principles of CRISPR Cas Enzymes Used in Nucleic Acid Detection,” Journal of Structural Biology 214, no. 1 (2022): 107838 DOI:1016/j.jsb.2022.107838
CrossRef - Hosen A, Nishat MNH, Soaib MMH, Sharkar OS, Sahabuddin M, Sharif IH, Bhajan SK. A review: CRISPR-Cas system and the mechanism with an inhibition of binding of CRISPR-Cas9. 2025; 6:e202400009 DOI:10.1002/nano.202400009
CrossRef - Khan FA, Pandupuspitasari NS, ChunJie H, Ahmad HI, Wang K, Ahmad MJ, Zhang S. Applications of CRISPR/Cas9 in reproductive biology. Curr Issues Mol Biol. 2017; Chapter 8. doi:10.21775/9781910190630.08.
CrossRef - Uludağ H, Aliabadi HM, Gasiunas G. Editorial: Current approaches to CRISPR/Cas9 delivery. Front Bioeng Biotechnol. 2022;10:1103007. doi: 10.3389/fbioe.2022.1103007
CrossRef - Onuma A, Fujii W, Sugiura K, Naito K. Efficient mutagenesis by CRISPR/Cas system during meiotic maturation of porcine oocytes. J Reprod Dev. 2017 Feb 16;63(1):45-50. doi: 10.1262/jrd.2016-094. Epub 2016 Oct 21. PMID: 27773884; PMCID: PMC5320429.
CrossRef - Wang HQ, Wang T, Gao F, Ren WZ. Application of CRISPR/Cas Technology in Spermatogenesis Research and Male Infertility Treatment. Genes (Basel). 2022 Jun 1;13(6):1000. doi: 10.3390/genes13061000. PMID: 35741761; PMCID: PMC9223233.
CrossRef - Bai M, Liang D, Wang Y, Li Q, Wu Y, Li J. Spermatogenic cell-specific gene mutation in mice via CRISPR-Cas9. J Genet Genomics. 2016;43(6):429. doi: 10.1016/j.jgg.2016.02.003. https://doi.org/10.1016/j.jgg.2016.02.003
CrossRef - Kosova G, Scott NM, Niederberger C, Prins GS, Ober C. Genome-wide association study identifies candidate genes for male fertility traits in humans. Am J Hum Genet. 2012 Jun 8;90(6):950-61. doi: 10.1016/j.ajhg.2012.04.016. Epub 2012 May 24. PMID: 22633400; PMCID: PMC3370277.
CrossRef - Wu Y, Liang D, Wang Y, Bai M, Tang W, Bao S, Yan Z, Li D, Li J. Correction of a genetic disease in mouse via use of CRISPR-Cas9. Cell Stem Cell. 2013 Dec 5;13(6):659-62. doi: 10.1016/j.stem.2013.10.016. PMID: 24315440.
CrossRef - Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014 Jun 5;157(6):1262-1278. doi: 10.1016/j.cell.2014.05.010. PMID: 24906146; PMCID: PMC4343198.
CrossRef - Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. 2014 Nov 28;346(6213):1258096. doi: 10.1126/science.1258096. PMID: 25430774.
CrossRef - Wu, Y., Zhou, H., Fan, X. et al.Correction of a genetic disease by CRISPR-Cas9-mediated gene editing in mouse spermatogonial stem cells. Cell Res 25, 67–79 (2015). https://doi.org/10.1038/cr.2014.160
CrossRef - Zhang YR, Yin TL, Zhou LQ. CRISPR/Cas9 technology: applications in oocytes and early embryos. J Transl Med. 2023 Oct 24;21(1):746. doi: 10.1186/s12967-023-04610-9. PMID: 37875936; PMCID: PMC10594749.
CrossRef - Wei Y, Lang J, Zhang Q, Yang CR, Zhao ZA, Zhang Y, Du Y, Sun Y. DNA methylation analysis and editing in single mammalian oocytes. Proc Natl Acad Sci U S A. 2019 May 14;116(20):9883-9892. doi: 10.1073/pnas.1817703116. Epub 2019 Apr 22. PMID: 31010926; PMCID: PMC6525536.
CrossRef - Wei Y, Yang CR, Zhao ZA. Viable offspring derived from single unfertilized mammalian oocytes. Proc Natl Acad Sci U S A. 2022 Mar 22;119(12):e2115248119. doi: 10.1073/pnas.2115248119. Epub 2022 Mar 7. PMID: 35254875; PMCID: PMC8944925.
CrossRef - Liu XS, Wu H, Ji X, Stelzer Y, Wu X, Czauderna S, Shu J, Dadon D, Young RA, Jaenisch R. Editing DNA Methylation in the Mammalian Genome. 2016 Sep 22;167(1):233-247.e17. doi: 10.1016/j.cell.2016.08.056. PMID: 27662091; PMCID: PMC5062609.
CrossRef - Suzuki T, Asami M, Perry AC. Asymmetric parental genome engineering by Cas9 during mouse meiotic exit. Sci Rep. 2014 Dec 23;4:7621. doi: 10.1038/srep07621.
CrossRef - Hashimoto M, Yamashita Y, Takemoto T. Electroporation of Cas9 protein/sgRNA into early pronuclear zygotes generates non-mosaic mutants in the mouse. Dev Biol. 2016 Oct 1;418(1):1-9. doi: 10.1016/j.ydbio.2016.07.017. Epub 2016 Jul 26. PMID: 27474397.
CrossRef - Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013 May 9;153(4):910-8. doi: 10.1016/j.cell.2013.04.025. Epub 2013 May 2. PMID: 23643243; PMCID: PMC3969854.
CrossRef - Fujii W, Kawasaki K, Sugiura K, Naito K. Efficient generation of large-scale genome-modified mice using gRNA and CAS9 endonuclease. Nucleic Acids Res. 2013 Nov;41(20):e187. doi: 10.1093/nar/gkt772. Epub 2013 Aug 30. PMID: 23997119; PMCID: PMC3814358.
CrossRef - Fujii W, Onuma A, Sugiura K, Naito K. Efficient generation of genome-modified mice via offset-nicking by CRISPR/Cas system. Biochem Biophys Res Commun. 2014 Mar 21;445(4):791-4. doi: 10.1016/j.bbrc.2014.01.141. Epub 2014 Jan 31. PMID: 24491566.
CrossRef - Fujii W, Onuma A, Sugiura K, Naito K. One-step generation of phenotype-expressing triple-knockout mice with heritable mutated alleles by the CRISPR/Cas9 system. J Reprod Dev. 2014;60(4):324-7. doi: 10.1262/jrd.2013-139. Epub 2014 May 4. PMID: 25110137; PMCID: PMC4139508.
CrossRef - Whitworth KM, Lee K, Benne JA, Beaton BP, Spate LD, Murphy SL, Samuel MS, Mao J, O’Gorman C, Walters EM, Murphy CN, Driver J, Mileham A, McLaren D, Wells KD, Prather RS. Use of the CRISPR/Cas9 system to produce genetically engineered pigs from in vitro-derived oocytes and embryos. Biol Reprod. 2014 Sep;91(3):78. doi: 10.1095/biolreprod.114.121723. Epub 2014 Aug 6. PMID: 25100712; PMCID: PMC4435063.
CrossRef - Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013 Feb 15;339(6121):819-23. doi: 10.1126/science.1231143. Epub 2013 Jan 3. PMID: 23287718; PMCID: PMC3795411.
CrossRef - Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013 Feb 15;339(6121):823-6. doi: 10.1126/science.1232033. Epub 2013 Jan 3. PMID: 23287722; PMCID: PMC3712628.
CrossRef - Adikusuma F, Piltz S, Corbett MA, Turvey M, McColl SR, Helbig KJ, Beard MR, Hughes J, Pomerantz RT, Thomas PQ. Large deletions induced by Cas9 cleavage. Nature. 2018 Aug;560(7717): E8-E9. doi: 10.1038/s41586-018-0380-z. Epub 2018 Aug 8. PMID: 30089922.
CrossRef - Egli D, Zuccaro MV, Kosicki M, Church GM, Bradley A, Jasin M. Inter-homologue repair in fertilized human eggs? 2018 Aug;560(7717):E5-E7. doi: 10.1038/s41586-018-0379-5. Epub 2018 Aug 8. PMID: 30089924.
CrossRef - Ma, H., Marti-Gutierrez, N., Park, SW. et al.Ma et al. reply. Nature 560, E10–E23 (2018). https://doi.org/10.1038/s41586-018-0381-y
CrossRef - Schenkwein D, Ylä-Herttuala S. Gene Editing of Human Embryos with CRISPR/Cas9: Great Promise Coupled with Important Caveats. Mol Ther. 2018 Mar 7;26(3):659-660. doi: 10.1016/j.ymthe.2018.02.007. Epub 2018 Feb 22. PMID: 29477495; PMCID: PMC5911640.
CrossRef - Shiza H, Hamid N, Ali A, U A K Saddozai, M B Khawar, X Ji. Advancing CRISPR technologies in reproductive biology. Gene & Protein in Disease 2024, 3(1), 2701. https://doi.org/10.36922/gpd.2701
CrossRef - Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013 May 9;153(4):910-8. doi: 10.1016/j.cell.2013.04.025. Epub 2013 May 2. PMID: 23643243; PMCID: PMC3969854.
CrossRef - Archambeault DR, Matzuk MM. Disrupting the male germ line to find infertility and contraception targets. Ann Endocrinol (Paris). 2014 May;75(2):101-8. doi: 10.1016/j.ando.2014.04.006. Epub 2014 Apr 30. PMID: 24793995.
CrossRef - Yan C, Wang P, DeMayo J, DeMayo FJ, Elvin JA, Carino C, Prasad SV, Skinner SS, Dunbar BS, Dube JL, Celeste AJ, Matzuk MM. Synergistic roles of bone morphogenetic protein 15 and growth differentiation factor 9 in ovarian function. Mol Endocrinol. 2001 Jun;15(6):854-66. doi: 10.1210/mend.15.6.0662. PMID: 11376106.
CrossRef - Young SA, Aitken RJ, Ikawa M. Advantages of using the CRISPR/Cas9 system of genome editing to investigate male reproductive mechanisms using mouse models. Asian J Androl. 2015 Jul-Aug;17(4):623-7. doi: 10.4103/1008-682X.153851. PMID: 25994645; PMCID: PMC4492054.
CrossRef - Zhang XH, Tee LY, Wang XG, Huang QS, Yang SH. Off-target Effects in CRISPR/Cas9-mediated Genome Engineering. Mol Ther Nucleic Acids. 2015 Nov 17;4(11):e264. doi: 10.1038/mtna.2015.37. PMID: 26575098; PMCID: PMC4877446.
CrossRef - Yu Y, Leete TC, Born DA, Young L, Barrera LA, Lee SJ, Rees HA, Ciaramella G, Gaudelli NM. Cytosine base editors with minimized unguided DNA and RNA off-target events and high on-target activity. Nat Commun. 2020 Apr 28;11(1):2052. doi: 10.1038/s41467-020-15887-5. PMID: 32345976; PMCID: PMC7189382.
CrossRef - Bravo JPK, Liu MS, Hibshman GN, Dangerfield TL, Jung K, McCool RS, Johnson KA, Taylor DW. Structural basis for mismatch surveillance by CRISPR-Cas9. Nature. 2022 Mar;603(7900):343-347. doi: 10.1038/s41586-022-04470-1. Epub 2022 Mar 2.
CrossRef - Jeon J, Park JS, Min B, Chung SK, Kim MK, Kang YK. Retroelement Insertion in a CRISPR/Cas9 Editing Site in the Early Embryo Intensifies Genetic Mosaicism. Front Cell Dev Biol. 2019 Nov 8;7:273. doi: 10.3389/fcell.2019.00273. PMID: 31781562; PMCID: PMC6857330.
CrossRef - Rossant J. Gene editing in human development: ethical concerns and practical applications. 2018 Jul 25;145(16):dev150888. doi: 10.1242/dev.150888. PMID: 30045910.
CrossRef - Baylis F, Darnovsky M, Hasson K, Krahn TM. Human Germ Line and Heritable Genome Editing: The Global Policy Landscape. CRISPR J. 2020 Oct;3(5):365-377. doi: 10.1089/crispr.2020.0082. Erratum in: CRISPR J. 2021 Apr;4(2):301-302. doi: 10.1089/crispr.2020.0082.correx. PMID: 33095042.
CrossRef - Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan LW, Levy JM, Chen PJ, Wilson C, Newby GA, Raguram A, Liu DR. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019 Dec;576(7785):149-157. doi: 10.1038/s41586-019-1711-4. Epub 2019 Oct 21. PMID: 31634902; PMCID: PMC6907074.
CrossRef - The Royal Society; National Academy of Sciences; National Academy of Medicine; International Commission on the Clinical Use of Human Germline Genome Editing. Heritable Human Genome Editing. Washington (DC): National Academies Press (US); 2020 Sep 3. PMID: 32897669.
- The Lancet. CRISPR-Cas9: a world first? Lancet. 2018 Dec 8;392(10163):2413. doi: 10.1016/S0140-6736(18)33111-8. PMID: 30527398.
CrossRef
Abbrevations List
ART – Assisted reproductive technology
Cas9 – CRISPR-associated protein 9
CRISPR – Clustered Regularly Interspaced Short Palindromic Repeats
cRNA – Coding RNA
crRNA – CRISPR RNA
DNA – Deoxyribonucleic acid
DNMT – DNA Methyltransferase
DSB – Double strand break
gRNA – guide RNA
GV – Germinal Vesicle
HCM – Hypertrophic Cardiomyopathy
HDR – Homology-Directed Repair
HNH – Histidine-Asparagine-Histidine
IVF – In vitro fertilisation
MYBPC3 – Myosin-Binding Protein C, cardiac-type gene,
NHEJ – Non-Homologous End Joining
NOA – Non-Obstructive Azoospermia
NUC – Nuclease lobe
OTEs – Off-Target Effects
PAM – Protospacer Adjacent Motif
PGC – Primordial Germ Cells
PGD – Preimplantation genetic diagnosis
REC – Recognition lobe
RNA – Ribonucleic acid
RuvC – Resolvase domain C
sgRNA – Single Guide RNA
SSCs – Spermatogonial Stem Cells
ssODNs – Single-Stranded Oligodeoxynucleotides
TET – Ten-eleven translocation enzyme
tracrRNA – trans-activating CRISPR RNA.

This work is licensed under a Creative Commons Attribution 4.0 International License.





