Manuscript accepted on : 25-09-2025
Published online on: 30-09-2025
Plagiarism Check: Yes
Reviewed by: Dr. Sonam Sneha
Second Review by: Dr. Makhabbah Jamilatun
Final Approval by: Dr. Wagih Ghannam
Proteomic and Biochemical Analysis of Maize Hybrids (Zea Mays L.) Induced by Salt Stress
Departnent of Biochemistry, University College of Science, Osmania University, Hyderabad, Telangana State, India.
Corresponding Author Email: e.nagendram@gmail.com
ABSTRACT: Due of maize's moderate sensitivity to salt stress, soil salinity poses a major danger to its global output. Developing solutions for enhanced performance in saline circumstances may be aided by researching how maize plants react to salt stress, resistance mechanisms, and management alternatives. We used maize hybrids (NK6240, M900Gold) (Zea mays L.) to check their yield concerning soil salt stress condition. Using 2D polyacrylamide gel electrophoresis, biochemical and protein patterns were examined following the application of salt stress to developing plants and the spots of interest were subjected to Mass spectrometry. As per our research outcomes among differentially expressed proteins, chosen seven spots of interest for comparative analysis and Among these three proteins are up -regulated and remaining four proteins are down-regulated. Thus concluded both maize hybrids given response and the effect of soil saline stress conditions of both varieties (NK6240, M900Gold).
KEYWORDS: IEF; MS; MALDI-TOF; Maize; Salt Stress
| Copy the following to cite this article: Nagendram E. Proteomic and Biochemical Analysis of Maize Hybrids (Zea Mays L.) Induced by Salt Stress. Biotech Res Asia 2025;22(3). |
| Copy the following to cite this URL: Nagendram E. Proteomic and Biochemical Analysis of Maize Hybrids (Zea Mays L.) Induced by Salt Stress. Biotech Res Asia 2025;22(3). Available from: https://bit.ly/3W3WNCd |
Introduction
One of the foremost critical cereal crops cultivated around the world is maize (Zea mays L.)and it positions third after wheat and rice generation. It is the most widely distributed crop with greater adaptability1. Around 790 million tons of maize are produced worldwide, and in certain nations, it is a staple grain that provides calories and proteins2 .The demand for maize will double in the developing world by 2050, and by 2025, it will be the most important crop produced both globally and in the developing world.3,4
Salinity is the most important abiotic stressor that prevents crop growth and productivity. Salt stress adversely affects plants’ functioning and metabolism and significantly hinders productivity5. Diffusion adjustment of halophytes and glycophytes is accomplished by improving organic and inorganic solutes. Therefore, a more significant decrease in cell substance potential than the external salt concentration could indicate a diffusion adjustment. Organic solutes are accumulated within the cytoplasm to balance the solute potential of the cavity, which is dominated by ions. It is noticed that the germination and seed plant stage of vegetation cycle is sensitive to salinity than the adult stage6. Plants response to salinity is one the foremost wide researched subjects in plant physiology7. Salinity affects plants in several ways diffusion affects specific particle toxicity and nutritionary disorders. It does not solely affect the morphology; however, it additionally modifies the metabolism of plants by limiting their growth.
The impact of proteins can be seen by observing the physiological responses to both osmotic and ionic effects of salinity. Considering the osmotic effect which causes not only a significant osmotic but also mechanical stress on plant cells, an enhanced biosynthesis of several somatically active organic compounds as well as proteins with osmoprotective functions such as LEA(late embryogenesis abundant) proteins could be mentioned.8
Proteins are expected to be significantly impacted by how plants react to environmental stresses such soil salinity. Large-scale changes in stress conditions will result in the identification of proteins and their corresponding genes that are concerned with the physiology of salt resistance. Utilizing 2D and mass spectrometry of controlled proteins whose blend was modified by salt treatment, a tall determination is gotten.
This work aimed to carry out proteomic analysis of the genes related to find out if they are tolerant or sensitive to salt stress. The purpose of this study was to learn more about the impact of salt stress and proteomic analysis in maize hybrids (Zea maize. L).This work pointed to carry out proteomic investigation of the qualities related to discover out on the off chance that they are tolerant or delicate to salt stretch. The reason of this consider was to learn more almost the affect of salt stretch and proteomic investigation in maize half breeds (Zea maize. L).
Materials and Methods
Conditions for plant growth and material
The popular Maize hybrids NK6240, M900Gold, were used for study as these hybrids are widely accepted by farmers due to high yield and stability. After five minutes of surface sterilization with a 1% sodium hypochlorite solution, the seeds were rinsed with distilled water. Surface sterilized seeds were pre-soaked in Petri plates with different levels of salinity (50mM, 100mM, 150mM of NaCl) for 40 min taking control with distilled water. We kept the treated seeds on wet Whatman no. In acid-washed Petri dishes, place one filter paper and incubated in dark at 27°C overnight. The next day, they are transferred to pots filled with acid-washed sand. The plants were cultivated in a greenhouse with natural light conditions, which included air temperatures between 27°C and 35°C, light intensities between 450 and 500 mmol/m2/s, and a relative humidity of 75%9.
Each pot containing five plants was supplied with 20mL of water and N:P:K (10:10:10) nutrient solution on alternate days. Harvested two weeks after germination, the plants were dried in a thermally ventilated oven at 70°C until they reached a consistent mass for dry weight calculation. Standard procedures were followed in the calculation of growth parameters, including fresh weight, dry weight, number of roots, root length, shoot length, leaf surface area, and shoot length. Various biochemical analyses were conducted on leaf samples from maize plants that were two weeks old.
Growth analysis of plants
Plants were carefully uprooted after nine days and washed with distilled water. Shoot and root length were measured with the help of scale. Plant fresh weight was noted by electronic balance; one set of plants was taken for 2D analysis, germination, and biochemical parameters analysis; another set was kept in a hot air oven at 700C. Dry weights of plants were calculated with the help of electronic weighing balance after 4 days of incubation in a hot air oven. Plant-1 g is used to represent both fresh and dry weights.
2Dimentional Electrophoresis
Harvest and protein (sample) preparation for 2D Electrophoresis
After harvesting the plant material, a whole plant sample was gathered and homogenized using liquid nitrogen. The fresh and dry weight of the complete leaf material is determined. The tissue was ground under liquid nitrogen to break up the leaf material. Since lowering the temperature of the cell material inhibits the activity of the protease, all stages of the protein extraction process were conducted at 4˚C. The addition of metabolite extraction buffer (MEB) is followed by five minutes of homogenization. After 20 minutes of centrifuging the solution (Eppendorf 5810) at 4˚C, the supernatant was gathered and placed in a separate tube. The use of MEB such as Methanol, Chloroform and Water contents is to remove metabolites from protein sample. To the protein pellet, SDS buffer (Sodium dodecyl sulphate, Dithiothreitol, Tris) supplemented by protease inhibitor cocktail and PMSF (phenylmethylsulfonyl fluoride) was added. For 1g of tissue, 5mL of SDS buffer was added. After vortexing, the samples were incubated for 1hr on Gel rocker (Genie). Centrifugation was carried out for 15 minutes at 4˚C and 10,000 rpm. Separate the pellet (which was kept at -80°C) and add equal amounts of Tris-buffered Phenol (Sigma) to the supernatant (SDS buffer). Shake for 30 minutes at room temperature. At 4˚C, centrifuged for 30 minutes at 10,000xg.
Six volumes of a 100 mM ammonium acetate/methanol solution were added to the lower phenol layer. Centrifuge at 10,000xg for 15 minutes at 4˚C after incubating overnight at -20˚C. After removing the supernatant, wash the pellet in acetone that has been chilled beforehand and centrifuge it for 15 minutes at 4˚C at 10,000 rpm. This process was carried out twice. The pellet should be allowed to air dry before being dissolved in rehydration buffer (7M urea, 2M thiourea, 0–5 percent pharmalyte buffer (v/v, pH 3–10); 4 percent CHAPS; 30mM DTT; 20mM Tris–base, pH 8.8). 0.3mL of rehydration buffer was added to the pellet for solubilization of proteins. From each sample 10µL was loaded on to the 1D gel for normalization of the concentrations.
Isoelectric focusing (IEF) and 2D PAGE
50 µL of the sample was diluted with 250µL of rehydration buffer. Each sample was loaded on to the isoelectric focusing strip 4-7pH gradient Linear (GE), 18cm for rehydration of the samples by applying the following conditions: 10hrs rehydration; Temperature -20°C. It was done in an IPGphor chamber (GE) to focus the strips isoelectrically. Before the gels were put in the IEF cell, mineral oil was applied on top of them. Rapid ramping of the voltage from 250V to 10,000V was used, and the current was 45mA per strip until 70000V was reached. Following the completion of the first dimension, the strips were submerged in equilibration buffer (50 mM Tris–HCl, pH 8.8; 6M urea; 30% glycerol; 2% (w/v) SDS; bromophenol blue, 0.001 percent (w/v) containing 1% DTT (w/v)) and gently shaken for one hour. The strips were then incubated for a further forty-five minutes with slow stirring in equilibration buffer containing four percent (w/v) iodoacetamide without DTT. The strips were washed several times with SDS-PAGE running buffer (25mM Tris–base; 192mM glycine; 0.1% (w/v) SDS). The second dimension was obtained with 10% SDS gels. The gel was loaded with a molecular weight standard (Biorad) of 10 to 250 kDa in 10 to 250 kDa.
The basic side (pH-7) of the gel was where marker lane was placed. Marker dyes and strips were applied to the gel surface, and the mixture was sealed with 1% (w/v) agarose that contained 0.01% (w/v) bromophenol blue. The second dimension was done in a vertical gel electrophoresis chamber (Ettan Dalt Six unit) at 25°C with a steady current of 45mA per gel. When the bromophenol blue departed the gel, the electrophoresis was terminated. After being taken out, the gels were left in a fixative containing 50% methanol and 10% acetic acid for the entire night.
Staining and Image Analysis
The gels were submerged in a 0–0.02% sodium thiosulfate solution for two hours with three changes, and then they were briefly rinsed with water. Following that, the gels were incubated for one hour in a solution containing 0–2% silver nitrate. A 2 percent sodium carbonate solution was used to develop the gels following a quick water wash. Following the cessation of the reaction, the gels were preserved in 10% acetic acid.
2D Analysis
Gels were digitized by scanning on Epson XL 11000 with 300 dpi and computer-assisted 2D analysis The protocol was performed using Image Master 2D Platinum software version 7.0.6 (GE Healthcare). The total number of spots on the gels is calculated using the software and the spots that are differentially expressed between the two samples are identified.
Software Parameters
The area of interest was chosen by cropping the gels and the spots were detected using parameters like smooth with a limit of 2, a minimum area with a limit of 5 and saliency with a limit of 5. After spot detection, every spot is checked manually for a real spot since the software detects clouds of dust, artefacts, which need to be removed from the analysis. The spots are then manually edited using options like create, delete, split, merge, grow and shrink. The spots that are located at precisely the same spot on each gel are used as landmarks. The landmarked spots aid in precisely matching the gels. Following the completion of the gel matching, the data analysis is finished. All spots in the gels are automatically matched by the software, which also assigns spot ID to every spot in the gel set and match ID to the spots that match in the gels.
The protein spots can be represented in the form of 3D with the peak height denoted as its intensity. The program annotates all of the remaining spots after determining the molecular weight and pI for a small number of spots on the corresponding gels. The data obtained after matching the gels were stored in the form of Spot Table, Gel Table, Scatter Plot, Gel Analysis Table, Match Statistics Table, Annotation Table etc.
To analyse differential expression investigation, the coordinated spots of the treated gel are differentiated with those of the control. The differentially communicated (overlap alter) spots between the gels are gotten by taking the spot percent volumes. A overlap alter can be calculated by separating the rate volumes of treated spots by the rate volumes of control spots with the same coordinate ID. This proportion shows that all of these spots are over-expressed in the event that it is more prominent than 1.5, and under-expressed in the event that it is less than 0.5. Special protein spots that are as it were found in that gel will be display within the spots that did not coordinate in both gels. In 2D gels, each spot speaks to a protein or polypeptide, either with or without a post-transcriptional adjustment.
In-gel Digestion and Mass Spectrometry
Excised gel spots were cut into pieces and taken into neatly labelled Eppendorf. Gel pieces were first washed with MS Grade water and then destained using 15mM K3 [Fe (CN) 6] and 50mM Hypo in 1:1 ratio. Buffer washes were done using 25mM ammonium bicarbonate, pH 8.5, in MS Grade water, then 50% acetonitrile in the same buffer, the gel plugs were then dehydrated using 100% acetonitrile. Sequencing-grade (Promega) trypsin was used to digest the gel plugs, which were then reduced with 100 mM dithiothreitol, alkylated with 250 mM iodoacetamide, and incubated in 25 mM ammonium bicarbonate for the entire night at 37˚C. The peptides were collected in an Eppendorf after being extracted three times using 0–1 percent trifluoroacetic acid (v/v) in 50 percent acetonitrile (v/v) following incubation. Following vacuum drying, the extracted peptides were redissolved in a 1:2 ratio of 0–1% (v/v) trifluoroacetic acid in 100% acetonitrile. The peptides that were extracted were combined with HCCA (α-Cyano-4-hydroxycinnamic acid) matrix in a 1:1 ratio (5 mg/mL α-Cyano-4-hydroxycinnamic acid) in a 1:2 ratio of 0–1 percent TFA and 100 percent ACN. The resulting 2µL was spotted onto the MALDI plate [(MTP 384 ground steel (Bruker Daltonics, Germany)]. Following air drying, the sample was examined using the MALDI TOF/TOF ultra flex III instrument.
Results
Germination
Salt stress adversely affects the growth of the maize plants though the germination rate was quite appreciable. The NK6240 and M900Gold varieties of maize were induced by different salt concentration ranging from 50-150mM NaCl and their rate of germination was found to be decreased with increasing salt concentration due to alleviated osmotic pressure (Graph 1).
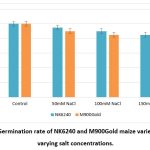 |
Graph 1: Germination rate of NK6240 and M900Gold maize varieties under varying salt concentrations. |
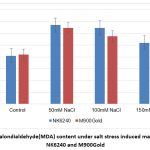 |
Graph 2: Malondialdehyde(MDA) content under salt stress induced maize varieties NK6240 and M900Gold |
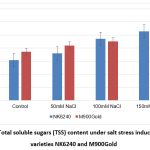 |
Graph 3: Total soluble sugars (TSS) content under salt stress induced maize varieties NK6240 and M900Gold |
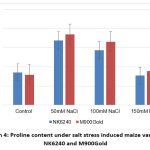 |
Graph 4: Proline content under salt stress induced maize varieties NK6240 and M900Gold |
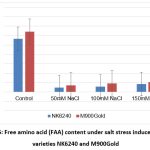 |
Graph 5: Free amino acid (FAA) content under salt stress induced maize varieties NK6240 and M900Gold |
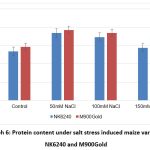 |
Graph 6 : Protein content under salt stress induced maize varieties NK6240 and M900Gold |
Table 1: Legend for table 1: Mean values of NK6240 and M900Gold for various biochemical parameters.
| NK 6240 | M900Gold | ||||||||
| Control | 50 Mm NaCl | 100 Mm NaCl | 150 Mm NaCl | Control | 50 Mm NaCl | 100 Mm NaCl | 150 Mm NaCl | ||
| Germination | 100 | 95 | 90 | 85 | 100 | 90 | 85 | 85 | |
| MDA | 8.2 | 13.4 | 12.9 | 10.4 | 8.4 | 12.9 | 11.5 | 10.2 | |
| TSS | 153.6 | 180.2 | 235.8 | 263.8 | 185.6 | 210 | 225.6 | 252.8 | |
| Proline | 8.56 | 16.9 | 14.4 | 7.7 | 7.95 | 18.5 | 16.6 | 8.9 | |
| FAA | 0.283 | 0.024 | 0.028 | 0.042 | 0.32 | 0.035 | 0.046 | 0.053 | |
| Protein | 13.4 | 18.4 | 17.3 | 14.4 | 14.6 | 19.2 | 18.4 | 16.9 | |
MDA (Malondialdehyde);TSS(Total Soluble Sugars);FAA(Free amino acids)
Biochemical Analysis
The various biochemical parameters such as malonialdehyde (MDA), total soluble sugars (TSS), Proline content, Free amino acids (FAA) and protein content of germinated maize plants of salt stress are discussed below in detail.
Malondialdehyde (MDA)
The trienoic fatty acids such as MDA directly contribute to reactive oxygen species (ROS) control via non-enzymatic oxidation and is directly correlated with the survival of tissues10. The maize plants have shown decreasing trend with increasing salt concentration, exhibiting the adaptable nature of the varieties NK6240 and M900Gold (Graph 2).
Total Soluble Sugars (TSS)
The elevated levels of soluble sugars contribute to tolerance of plants against stress conditions11. Similarly, the maize plants have shown increasing levels of TSS with increasing salt concentration (Graph 3). The maize variety M900Gold have shown high levels of TSS than NK6240, indicating its sensitivity against salt stress.
Proline
Proline acts as a osmoregulatory and protects plant proteins against damage and enhances various enzyme activities.12 Studies indicate that the elevated levels of Proline maintain NADP+/NADPH ratio. The maize varieties have shown elevated levels of Proline initially and then reduction under high salt concentration (Graph 4).
Free amino acids (FAA) and Protein
The free amino acids content was significant very less when compared with the control but have shown increasing trend in a dose dependent manner. Under stress conditions, the physiological process is significantly affected which can be correlated by the elevated protein levels.13 All the treated plants have shown elevated protein levels than control (Graph 5, Graph 6). Moreover, the treated have shown decreasing trend with increasing salt concentrations indicating its adaptive nature with high salt concentration.
Proteomic Analysis
Three up-regulated and four down-regulated proteins were among the seven proteins that were selected for additional investigation following proteomic analysis using 2D electrophoresis and MALDI-TOF-MS.(Table 2).
 |
Figure 1: Representation of 2D gel electrophoresis images of maize hybrids (a) GC control gel and (b) GT treated with NaCl. |
Table 2: Up-regulated and Down-regulated proteins expressed in two dimentional electrophoresis.
| MATCH ID | SPOT ID | Uniport Accession Number | Name of the Protein |
| Down regulated proteins expressed in salt induced stress | |||
| 18 | 1168 | Q995P4 | CRS2_MAIZE Chloroplastic group IIB intron splicing Facilitator CRS 2 |
| 101 | 1050 | P24631 | HSP 21_MAIZE 17.5kDa
class II heat shock protein |
| 14 | 1176 | Q4G2J5 | DER12_MAIZE Derlin-1.2 |
| 127 | 991 | Q41793 | CDPK_MAIZE Calcium –dependent protein kinase |
| Up-regulated proteins expressed in salt induced stress | |||
| 996 | – | D9J101 | OMT8_MAIZE Benzoate O-methyltransferase |
| 331 | 1147 | O81482 | IF4E2_MAIZE Eukaryotic translation initiation factor |
| 41 | 1134 | Q43266 | PCNA_MAIZE Proliferating cell nuclear antigen |
Functions of up-regulated proteins
OMT8_MAIZE Benzoate O-methyltransferase
In reaction to stress, methyltransferases play a role in the biosynthesis of methyl benzoate.. These transferases utilize exclusively benzoic acid as a substrate. In our research study, we induced saline stress, so that this protein might be up-regulated. An anthranilic acid methyltransferase-one (AAMT1) appears to be responsible for most of the activity of S-adenosyl-L-methionine-dependent methyltransferase and the formation of methyl anthranilate observed in maize after harm to herbivores. The enzymes may also be involved in the formation of low amounts of methyl salicylate, which are emitted from herbivore-damaged maize 14. The methylation of the carboxyl group of several low molecular weight metabolites is catalyzed by plant enzymes belonging to the SABATH methyltransferase family, which is essential to the plant’s life cycle.15,16.
IF4E2_MAIZE Eukaryotic translation initiation factor
Eukaryotic translation initiation factor participates in forming translation initiation factor 4f complex and proceed for translation. Beginning, elongating, and terminating are the three stages of the synthesis of mRNA proteins, which is a crucial step in the expression of eukaryotic genes. Acknowledges and attaches itself to the 7-methylguanosine that forms the mRNA cap at an early stage of protein synthesis and encourages ribosome binding by causing secondary structure mRNA unwinding. In our results, there is no salt stress effect in translation processes 17. Hence there is no effect of salt stress on this protein. Hence this protein is up-regulated.
PCNA_MAIZE proliferating cell nuclear antigen
This is an auxiliary protein of the DNA polymerase delta which is involved in regulating the replication of eukaryotic DNA by the process ability of the polymerase during elongation of the leading strand. The multipurpose protein PCNA is involved in the synthesis of DNA replication, DNA repair, and recombination-driven DNA. It belongs to the subunit of DNA polymerases d and e, which have both been linked to repair processes such as post-replication repair, NER, BER, mismatch repair, and recombination-driven DNA synthesis18. Additionally, proteins involved in non-homologous end-joining, homologous recombination, and other crucial DNA replication processes are bound by PCNA18. According to our results, there is no maximum salt stress effect on the role of protein hence it is up-regulated.
Down-Regulated Proteins
CRS2_MAIZE Chloroplastic group IIB intron splicing Facilitator CRS 2
This protein plays a role in splicing of group II B introns in chloroplasts. This chloroplast RNA splicing protein complex with either CAF1 or CAF2 which, in turn, interact with RNA and confer introns specificity of the splicing particles. CRS2 has no peptidyl t-RNA hydrolase activity19. According to our results, salt stress effects are shown on this protein hence this protein has been down-regulated. Maize hybrids growth is also altered when compared to control.
HSP 21_MAIZE (17.5kDa) class II heat shock protein
The main role of this protein is in protein complex oligomerization, protein folding, responding to heat, responding to high light intensity and also responding to hydrogen peroxide. We have applied salt stress in our experiment and all the above biological processes are affected, so this protein has been down-regulated 20. Due to this effect Maize hybrids, growth is also altered when compared to control.
DER12_MAIZE Derlin-1.2
This protein contributes to the breakdown of particular misfolded luminal proteins in the endoplasmic reticulum (ER). One crucial aspect of this protein quality control is the removal of misfolded proteins. Previous research using a range of soluble and transmembrane associated degradation (ERAD) substrates showed variations in the ER degradation machinery employed21. In our study, due to the induction of salt stress in maize hybrids, the role of protein derlin is denied, so this protein has been down-regulated. Also, growth levels of maize hybrids have been decreased due to salt stress when compared to control.
CDPK_MAIZE Calcium-dependent protein kinase
The main role of the protein calcium-dependent protein kinase in cell involves ATP binding mechanisms, calcium ion binding processes and protein serine /threonine activity controlled by the CDPK. Also, their functions include phosphorylation, which plays important role in plant calcium signal transduction and response to osmotic stresses22. Our results indicate that this protein has been down-regulated due to salt stress and also decrease in their growth levels of maize hybrids.
Discussion
In the case of gemination of maize varities Salt stress adversely affects the growth of the maize plants though the germination rate was quite appreciable. The NK6240 and M900Gold varieties of maize were induced by different salt concentration and their rate of germination was found to be decreased with increasing salt concentration due to alleviated osmotic pressure.Within the case of gemination of maize varities Salt push antagonistically influences the development of the maize plants in spite of the fact that the germination rate was very calculable. The NK6240 and M900Gold assortments of maize were actuated by diverse salt concentration and their rate of germination was found to be diminished with expanding salt concentration due to lightened osmotic weight.
In the case of Malondialdehyde (MDA) The trienoic fatty acids such as MDA directly contribute to reactive oxygen species (ROS) control via non-enzymatic oxidation and is directly correlated with the survival of tissues. The maize plants have shown decreasing trend with increasing salt concentration, exhibiting the adaptable nature of the varieties NK6240 and M900Gold.
Total Soluble Sugars (TSS)
The elevated levels of soluble sugars contribute to tolerance of plants against stress conditions. Similarly, the maize plants have shown increasing levels of TSS with increasing salt concentration . The maize variety M900Gold have shown high levels of TSS than NK6240, indicating its sensitivity against salt stress.
Proline
Proline acts as a osmoregulatory and protects plant proteins against damage and enhances various enzyme activities. Our research results indicate that the elevated levels of Proline maintain NADP+/NADPH ratio. The maize varieties have shown elevated levels of Proline initially and then reduction under high salt concentration.
Free amino acids (FAA) and Protein:
The free amino acids substance was critical exceptionally less when compared with the control but have appeared expanding slant in a measurements subordinate way decreasing conditions, the physiological prepare is altogether influenced which can be related by the elevated protein levels. All the treated plants have appeared hoisted protein levels than control. Additionally, the treated have appeared diminishing drift with expanding salt concentrations showing its versatile nature with enhanced salt concentration.
Four proteins down- regulated proteins expressed in salt induced stress which are including
(OMT8_MAIZE)BenzoateO-methyltransferaseBenzoateO-methyltransferase,
(IF4E2_MAIZE )Eukaryotic translation initiation factor,
(PCNA_MAIZE) Proliferating cell nuclear antigen
Four proteins down- regulated proteins expressed in salt induced stress which are including
CRS2_MAIZE Chloroplastic group IIB intron splicing Facilitator CRS
HSP 21_MAIZE 17.5kDa class II heat shock protein
DER12_MAIZE Derlin-1.2
CDPK_MAIZE Calcium –dependent protein kinase
Indicates the tolerance of maize hybrids (NK6240, M900Gold) to salt stress and is indicated by proteomic analysis. Also, our data demonstrated that salt stress reduces the deleterious effects of soil salinity and drought in the maize plant.
Summary
Our research results showed that salt stress lessens the negative effects of drought and soil salinity on maize plants. Abiotic stressors like drought and salinity affect maize yields by causing physiological and biochemical alterations like ionic imbalance and decreased photosynthesis. According to study findings, salt stress can lessen the negative effects of drought and soil salinity on maize plants. Finding hybrids that can withstand salt, such as NK6240 and M900Gold, is essential.
Conclusion
In the global economy and food industry, maize plays a significant role. In terms of production and cultivated area, it is currently ranked second in importance, after rice. Thus, it is crucial to preserve maize under abiotic stressors such as salt stress, UV stress, metal stress, and cold stress. Our findings, which were supported by proteomic analysis, demonstrated that the maize hybrids (NK6240, M900Gold) were resistant to salt stress.
Acknowlwdgements
Nagendram Erram would like to thank the Department of Biochemistry.
Funding Sources
This work was supported in part from the grant ,UGC-BSR-RFMS & UGC- to Professor. Manjula Bhanoori.
Conflict of Interest
The authors do not have any conflict of interest.
Data Availability Statement
This statement does not apply to this article.
Ethics Statement
This research did not involve human participants, animal subjects, or any material that requires ethical approval.
Informed Consent Statement
This study did not involve human participants, and therefore, informed consent was not required.
Clinical Trial Registration
This research does not involve any clinical trials.
Permission to reproduce material from other sources
Not Applicable
Author Contributions
The sole author was responsible for the conceptualization, methodology, data collection, analysis, writing, and final approval of the manuscript
References
- Alamer KH, Perveen S, Khaliq A,et al. Mitigation of Salinity Stress in Maize Seedlings by the Application of Vermicompost and Sorghum Water Extracts. Plants (Basel). 2022 Sep 28;11(19):2548. doi: 3390/plants11192548. PMID: 36235413; PMCID: PMC9572175.
CrossRef - Ambikapathy J, Marshall JS, Hocart CH, Hardham AR. The role of proline inosmoregulation in Phytophthora nicotianae. Fungal Genet Biol. 2002Apr;35(3):287-99. doi: 10.1006/fgbi.2001.1327. PMID: 11929217.
CrossRef - 3.Shiferaw B, Prasanna BM, Hellin J, et al. Crops that feed the world 6. Past successes and future challenges to the role played by maize in global food security. Food security. 2011 Sep;3(3):307-27. DOI:10.1007/s12571-011-0140-5
CrossRef
- Fonseca, J., Narrod, C., Rosegrant, M.W.et al. Looking into the future for agriculture and AKST. https://cgspace.cgiar.org/items/897ffb3a-23d9-41e7-ad3e- 4b2546614c17
CrossRef - Shao, J. et al. Versatile roles of polyamines in improving abiotic stress tolerance of plants, Frontiers. 2025. DOI:3389/fpls.2022.1003155
CrossRef - Wang Z, Großkinsky DK, Li D, Zheng W. Editorial: Physiological and molecular mechanisms of important agronomic traits in plants under various abiotic factors. FrontPlantSci. 2024 Oct18;15:1502061. doi:10.3389/fpls.2024.1502061. PMID: 39494063; PMCID:
CrossRef - Negrão S, Schmöckel SM, Tester M. Evaluating physiological responses of plants to salinity stress. Ann Bot. 2017 Jan;119(1):1-11. doi: 1093/aob/mcw191. Epub 2016 Oct 5. PMID: 27707746; PMCID: PMC5218372.
CrossRef - Bhardwaj R, Sharma I, Kanwar M, et al. LEA proteins in salt stress Salt stress in plants: Signalling, omics and adaptations. 2013 Jan 12:79-112. DOI:10.1007/978-1-4614-6108-1_5
CrossRef - Rady MM, Varma B, Howladar SM. et al. Common bean (Phaseolus vulgaris L.) seedlings overcome NaCl stress as a result of presoaking in Moringa oleifera leaf extract. Scientia Horticulturae. 2013 Oct 23;162:63-70. https://doi.org/10.1016/j.scienta.2013.07.046
CrossRef - Mene-Saffrane L, Dubugnon L, Chetelat A, et al.Nonenzymatic oxidation of trienoic fatty acids contributes to reactive oxygen species management in Journal of Biological Chemistry. 2009 Jan 16;284(3):1702-8. DOI 10.1074/jbc.M807114200
CrossRef - Correia CM, Areal EL, Torres-Pereira MS, et al. Intraspecific variation in sensitivity to ultraviolet-B radiation in maize grown under field conditions: II. Physiological and biochemical aspects. Field crops research. 1999 Jun 25;62(2-3):97-105. https://doi.org/10.1016/S0378-4290(98)00164-6
CrossRef - Khan P, Abdelbacki AMM, Albaqami M, et al. Proline Promotes Drought Tolerance in Maize. Biology (Basel). 2025 Jan 7;14(1):41. doi:10.3390/biology14010041. PMID: 39857272; PMCID:
CrossRef - Batista-Silva W, Heinemann B, Rugen N, et al.. The role of amino acid metabolism during abiotic stress release. Plant Cell Environ. 2019 May;42(5):1630-1644. doi: 10.1111/pce.13518. Epub 2019 Feb 7. PMID:
CrossRef - Köllner TG, Lenk C, Zhao N, et al. Herbivore-induced SABATH methyltransferases of maize that methylate anthranilic acid using S- adenosyl-L-methionine. Plant physiology. 2010 Aug 1;153(4):1795-807. https://doi.org/10.1104/pp.110.158360
CrossRef - Zhuge XL, Du X, Xiu ZJ, et al. Discovery of specific catalytic activity toward IAA/FA by LaSABATHs based on genome- wide phylogenetic and enzymatic analysis of SABATH gene family from Larix kaempferi.Int J Biol Macromol. 2023 Jan 15;225:1562- doi: 10.1016/j.ijbiomac.2022.11.212. Epub 2022 Nov 25. PMID: 36442561.
CrossRef - Köllner TG, Lenk C, Zhao N, et al. Herbivore-induced SABATH methyltransferases of maize that methylate anthranilic acid using S- adenosyl-L-methionine.Plantphysiology.2010Aug1;153(4):1795-1807 https://doi.org/10.1104/pp.110.158360
CrossRef - Sesma A, Castresana C, Castellano MM. Regulation of Translation by TOR, eIF4E and eIF2α in Plants: Current Knowledge, Challenges and Future Front Plant Sci. 2017 Apr 26;8:644. doi: 10.3389/fpls.2017.00644. PMID: 28491073; PMCID: PMC5405063.
CrossRef - Paunesku T, Mittal S, Protić M, et al. Proliferating cell nuclear antigen (PCNA): ringmaster of the genome. Int J Radiat Biol. 2001 Oct;77(10):1007-21. doi: 1080/09553000110069335. PMID: 11682006.
CrossRef - del Campo EM. Post-transcriptional control of chloroplast gene expression. Gene Regul Syst Bio. 2009 Mar 12;3:31-47. doi: 10.4137/grsb.s2080. PMID:19838333; PMCID: PMC2758277.
CrossRef - Mas-ud MA, Juthee SA, Hosenuzzaman M, et al. Current Understanding of Heat Shock Protein-Mediated Responses to Heat Stress in Environmental and Experimental Botany.2025Jul5:106192. https://doi.org/10.1016/j.envexpbot.2025.106192
CrossRef - Taxis C, Hitt R, Park SH, et al. Use of modular substrates demonstrates mechanistic diversity and reveals differences in chaperone requirement of J Biol Chem. 2003 Sep 19;278(38):35903-13. doi:10.1074/jbc.M301080200. Epub 2003 Jul 7. PMID: 12847107.
CrossRef - Klimecka M, Muszyńska G. Structure and functions of plant calcium- dependent protein kinases. Acta Biochim Pol. 2007;54(2):219-33. Epub 2007 Apr 19. PMID: 17446936
CrossRef
Abbrevations List
MS = Mass Spectrometry
MALDI-TOF = Matrix-Assisted Laser Desorption/Ionization–Time-Of-Flight
LEA = Late embryogenesis abundant
MEB = Metabolic extraction buffer
SDS buffer = Sodium dodecyl sulphate, Dithiothreitol, Tris.
CHAPS = 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate.
DTT= Dithiothreitol
PMSF= phenylmethylsulfonyl fluoride
GE= gradient Linear
SDS-PAGE= sodium dodecyl sulphate -polyacrylamide gel electrophoresis
pI = Isoelectric point
HCCA = α-Cyano-4-hydroxycinnamic acid
TFA= trifluoroacetic acid
ROS= reactive oxygen species
ACN= acetonitrile
MDA = Malondialdehyde;
TSS = Total Soluble Sugars;
FAA = Free amino acids
OMT8_MAIZE = BenzoateO-methyltransferaseBenzoateO-methyltransferase,
IF4E2_MAIZE = Eukaryotic translation initiation factor,
PCNA_MAIZE = Proliferating cell nuclear antigen
CRS2_MAIZE = Chloroplastic group IIB intron splicing Facilitator
CRS HSP 21_MAIZE = 17.5kDa class II heat shock protein
DER12_MAIZE = Derlin-1.2
CDPK_MAIZE = Calcium –dependent protein kinase
AAMT1 = An anthranilic acid methyltransferase-one

This work is licensed under a Creative Commons Attribution 4.0 International License.





