Manuscript accepted on : 05-06-2025
Published online on: 16-06-2025
Plagiarism Check: Yes
Reviewed by: Dr. Satishkumar Khadia
Second Review by: Dr. Vivek Anand
Final Approval by: Dr. Wagih Ghannam
Fouzia Hilal and Jithesh Krishnan Ramakrishnan Nair*
and Jithesh Krishnan Ramakrishnan Nair*
Department of Botany, NSS College, Pandalam, Pathanamthitta district, Kerala.
Correspondence Author Email: kjith77@gmail.com
DOI : http://dx.doi.org/10.13005/bbra/3381
ABSTRACT: Dalzellia ubonensis M. Kato (2006), previously known only from Laos, Thailand, and Vietnam, is reported here for the first time from India, specifically from the Kallar River in the Vadasserikkara Range, Pathanamthitta district, Kerala. Detailed morphological examination revealed key diagnostic traits—absence of roots, broad crustose shoots, rosette leaf arrangement, and numerous ovules—that match the protologue of D. ubonensis and distinguish it from other Indian congeners, particularly Dalzellia gracilis. For molecular confirmation, genomic DNA was extracted and the plastid matK region was amplified and sequenced. BLASTn analysis showed ≥98% sequence identity and 100% query coverage with reference Dalzellia sequences in GenBank, corroborating its identity as D. ubonensis. This integrative taxonomic approach—combining classical morphological taxonomy with DNA barcoding—confirms a significant range extension for the species and represents the first verified record of D. ubonensis in India. This finding highlights the floristic uniqueness of Kerala’s riverine ecosystems and underscores the need for targeted exploration and conservation of aquatic Podostemaceae taxa in understudied regions of the Western Ghats.
KEYWORDS: Chloroplast; Dalzellia; Genome; Morphology; Phylogeny; Podostemaceae
Download this article as:| Copy the following to cite this article: Hilal F, Nair J. K. R. Dalzellia ubonensis (Podostemaceae): A New Record for India Evidenced by Morphological Characters and Phylogenetic Analysis. Biotech Res Asia 2025;22(2). |
| Copy the following to cite this URL: Hilal F, Nair J. K. R. Dalzellia ubonensis (Podostemaceae): A New Record for India Evidenced by Morphological Characters and Phylogenetic Analysis. Biotech Res Asia 2025;22(2). Available from: https://bit.ly/3FR3C5w |
Introduction
The Podostemaceae generally known as river-weeds are distinctive aquatic angiosperms found in wetlands throughout the tropics and subtropics worldwide.1-3 These plants thrive in fast-flowing, turbulent currents, firmly adhering to rock surfaces during the rainy season. As water levels recede during the dry season, they germinate, blossom, produce fruit, and eventually wither. During the rainy season, seeds dispersed by running water attach to rock surfaces, where they germinate and grow into seedlings.4 The subfamily Tristichoideae of Podostemaceae consists of approximately 20 species distributed across five genera globally, with Asia 5-7 being the centre of species diversity. The five genera in this subfamily are Terniopsis, Tristicha, Indodalzellia, Indotristicha, and Dalzellia.8-7 Out of these genera, the distribution map of Dalzellia 6 shows that Dalzellia ubonensis, D. ranongensis, D. kailarsenii, and D. angustissima occur in Thailand; and D. zeylanica and D. gracilis, in India and Sri Lanka, and India, respectively.
During floristic surveys conducted in the Kallar River (Latitude 9.240924o, Longitude 76.965226o), a tributary of the Pampa River in the Vadasserikkara Range of the Ranni Forest Division, Pathanamthitta District, Kerala, India, the authors collected several intriguing specimens of Dalzellia.
Taxonomic evaluation of the specimens, along with a review of relevant literature and molecular studies, revealed that the species collected from the aforementioned area corresponds to Dalzellia ubonensis M. Kato. Dalzellia gracilis, reported from India, is a closely related species to Dalzellia ubonensis. However, it differs from D. ubonensis in several key characteristics: the presence of roots, numerous root-borne shoots, and a crustose shoot that is irregularly shaped and nearly ribbon-like with hapters. Additionally, D. gracilis lacks the cupula at the base of the floriferous shoot and does not have “rosettes” on the upper side of the crust,6,11 which are present in D. ubonensis. Morphological studies, supported by literature citations and robust genomic analysis, confirm that this is the first report of Dalzellia ubonensis from the state of Kerala, India. The voucher specimens were deposited at Kerala University herbarium (KUBH11308) and at the Post Graduate and Research Department of Botany, NSS College, Pandalam (NSSPDMBODHOS7) respectively. A detailed account of the species is presented below.
Taxonomic Treatment
Dalzellia ubonensis M. Kato, Acta Phytotax. Geobot. 57(1): 10 (2006).
The shoot of Dalzellia ubonensis is crustose (foliose), slightly pinkish in color, and adheres to rocky substrates like a foliose lichen, with no roots present 10-11 (Fig.1B). The shoot measures 3–10 mm or wider and is lobed. It exhibits dorsiventral construction, with subdimorphic leaves (Fig.1A) arranged on the upper surface. The lower surface of the shoot, which attaches to the substrate, is devoid of leaves (Fig.1A).
Dorsal leaves
Arranged in branched longitudinal rows (Fig.1G), these leaves are densely fimbriate, linear-oblong, and smaller than the marginal leaves, measuring approximately 1 mm × 0.1 mm. They are rounded at the apex and oriented toward the distal end of the row.
Marginal (lateral) leaves
Narrowly deltoid, fimbriate, and rounded at the apex, these leaves are larger, reaching about 2 mm × 0.5 mm.
Rosette leaves
Scattered on the dorsal surface of the shoot, these leaves are linear, rounded at the apex, and measure approximately 2 mm × 0.2 mm.
Flowers are scattered on the shoots
Peduncle: 5–9 mm long (Fig.1C,1H).
Calyx: Membranous tepals as long as the ovary, shallowly 3-lobed, with deeper incisions at anthesis.
Stamens: Three, with di-thecous anthers, each filament 2 mm long, and equal brown (Fig.1H) lobes longer than the ovary.
Ovary: Obovoid-ellipsoid, about 2 mm long and 1 mm thick, trilocular with axile placentation; ovules number roughly 30 per locule.
Stigmas: Three (Fig.1C), papillate, 0.2–0.3 mm long.
Capsule: Stalked (10 mm long), dark brown, trigonous, measuring approximately 2 mm × 1 mm, with nine ribs. After dehiscence, the valves curve inward (Fig.1D), revealing numerous seeds, about 30 per locule (Fig.1E).
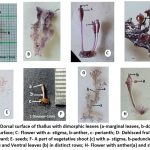 |
Figure 1: A- Dorsal surface of thallus with dimorphic leaves (a-marginal leaves, b-dorsal leaves); B- Ventral surface; C- Flower with a- stigma, b-anther, c- perianth; D- Dehisced fruit with valve refluxed inward;Click here to view Figure |
Phenology
We observed both flowers and fruits during January (in India).
Distribution: Thailand, Vietnam, Laos, India.
Phylogenetic Studies
DNA Extraction, Amplification and Sequencing
DNA Barcoding using universal primers of MATK
DNA isolation using NucleoSpin® Plant II Kit (Macherey-Nagel)
About 100 mg of the tissue/mycelium is homogenized using liquid nitrogen and the powdered tissue is transferred to a microcentrifuge tube. Four hundred microlitres of buffer PL1 is added and vortexed for 1 minute. Ten microlitres of RNase A solution is added and inverted to mix. The homogenate is incubated at 65oC for 10 minutes. The lysate is transferred to a Nucleospin filter and centrifuged at 11000 x g for 2 minutes. The flow through liquid is collected and the filter is discarded. Four hundred and fifty microlitres of buffer PC is added and mixed well. The solution is transferred to a Nucleospin Plant II column, centrifuged for 1 minute and the flow through liquid is discarded. Four hundred microlitre buffer PW1 is added to the column, centrifuged at 11000 x g for 1 minute and flow though liquid is discarded. Then 700 µl PW2 is added, centrifuged at 11000 x g and flow through liquid is discarded. Finally, 200 µl of PW2 is added and centrifuged at 11000 x g for 2 minutes to dry the silica membrane. The column is transferred to a new 1.7 ml tube and 50 µl of buffer PE is added and incubated at 65oC for 5 minutes. The column is then centrifuged at 11000 x g for 1 minute to elute the DNA. The eluted DNA was stored at 4oC.
Agarose Gel Electrophoresis for DNA Quality check
The quality of the DNA isolated was checked using agarose gel electrophoresis. 1µl of 6X gel-loading buffer (0.25% bromophenol blue, 30% sucrose in TE buffer pH-8.0) was added to 5µl of DNA. The samples were loaded to 0.8% agarose gel prepared in 0.5X TBE (Tris-Borate-EDTA) buffer containing 0.5 µg/ml ethidium bromide. Electrophoresis was performed with 0.5X TBE as electrophoresis buffer at 75 V until bromophenol dye front has migrated to the bottom of the gel. The gels were visualized in a UV transilluminator (Genei) and the image was captured under UV light using Gel documentation system (Bio-Rad).
PCR Analysis
Table 1: PCR Analysis
| 2X Phire Master Mix | 5μL |
| D/W | 4μL |
| Forward Primer | 0.25μL |
| Reverse Primer | 0.25μL |
| DNA | 1μL |
Primers used
Table 2: Primers used
| Target | Primer Name | Direction | Sequence (5’ à 3’) |
| MATK | MATK-XF | Forward | TAATTTACGATCAATTCATTC |
| MATK –NR1 | Reverse | ACAAGAAAGGCGAAGTAT |
The PCR amplification was carried out in a PCR thermal cycler (GeneAmp PCR System 9700, Applied Biosystems).
PCR amplification profile
MATK
98 oC – 30 sec
98 oC – 5 sec
45 oC – 10 sec }10 cycles
72 oC – 15 sec
98 oC – 5 sec
50 oC – 10 sec }30 cycles
72 oC – 15 sec
72 oC – 60 sec
4 oC – ∞
Agarose Gel electrophoresis of PCR products
The PCR products were checked in 1.2% agarose gels prepared in 0.5X TBE buffer containing 0.5 µg/ml ethidium bromide. 1 µl of 6X loading dye was mixed with 4 µl of PCR products and was loaded and electrophoresis was performed at 75V power supply with 0.5X TBE as electrophoresis buffer for about 1-2 hours, until the bromophenol blue front had migrated to almost the bottom of the gel. The molecular standard used was 2-log DNA ladder (NEB). The gels were visualized in a UV transilluminator (Genei) and the image was captured under UV light using Gel documentation system (Bio-Rad).
ExoSAP-IT Treatment
ExoSAP-IT (GE Healthcare) consists of two hydrolytic enzymes, Exonuclease I and Shrimp Alkaline Phosphatase (SAP), in a specially formulated buffer for the removal of unwanted primers and dNTPs from a PCR product mixture with no interference in downstream applications.
Five micro litres of PCR product is mixed with 0.5µl of ExoSAP-IT and incubated at 37oC for 15 minutes followed by enzyme inactivation at 85oC for 5 minutes.
Sequencing using BigDye Terminator v3.1
Sequencing reaction was done in a PCR thermal cycler (GeneAmp PCR System 9700, Applied Biosystems) using the BigDye Terminator v3.1 Cycle sequencing Kit (Applied Biosystems, USA) following manufactures protocol.
The Sequencing PCR mix consisted of the following components:
| D/W | 6.6μL |
| 5X Sequencing Buffer | 1.9μL |
| Forward Primer | 0.3μL |
| Reverse Primer | 0.3μL |
| Sequencing Mix | 0.2μL |
| Exosap treated PCR product | 1μL |
SequencingPCR amplification profile
96oC – 2min
96oC – 30sec
50oC – 40sec } 30 cycles
60 oC – 4min
4 oC – ∞
Post Sequencing PCR Clean up
| D/W | 5 µl |
| 3M Sodium Acetate | 1 µl |
| EDTA | 0.1 µl |
| 100% Ethanol | 44 µl |
Mix D/W, 125mM EDTA, 3M sodium acetate pH 4.6 and ethanol were prepared and were properly mixed.
50 µl of mix was added to each well in the sequencing plate containing sequencing PCR product.
Vortex by Mixmate vortex and incubated at room temperature for 30 minutes
Spun at 3700 rpm for 30 minutes
Decanted the supernatant and added 50 µl of 70% ethanol to each well
Spun at 3700 rpm for 20 minutes.
Decanted the supernatant and repeated 70% ethanol wash
Decanted the supernatant and air dried the pellet.
The cleaned-up air dried product was sequenced in ABI 3500 DNA Analyzer (Applied Biosystems).
Sequence Analysis
The sequence quality was checked using Sequence Scanner Software v1 (Applied Biosystems). Sequence alignment and required editing of the obtained sequences were carried out using Geneious Pro v5.112
Phylogenetic Analyses
The obtained DNA sequences were analyzed using the NCBI Nucleotide BLAST (BLASTn) tool (https://blast.ncbi.nlm.nih.gov/) to compare them with existing sequences in the GenBank database for species identification and similarity assessment. The sequences were submitted in FASTA format, and MEGA-BLAST, an algorithm optimized for highly similar sequences, was used for analysis. The results were filtered based on 100% query coverage and ≥98% sequence identity to ensure accurate species identification. The BLAST output provided a ranked list of matching sequences, displaying key parameters such as Max Score, E-value, Identity Percentage, and Query Coverage. The top hits were carefully examined, and sequences with high similarity consistently matched species within the genus Dalzellia (Fig.4). The strong alignment between the obtained sequences and the reference sequences in the database confirmed the taxonomic identity of the plant sample. This analysis provided molecular validation of the plant’s classification, demonstrating a high degree of confidence in its genetic identification.
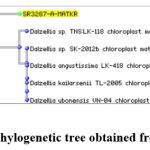 |
Figure 4: Phylogenetic tree obtained from BLASTClick here to view Figure |
Result
From the phylogenetic analysis the plant specimen collected from Kallar river (Vadasserikkara range) of Pathanamthitta district of Kerala, India was identified as belonging to Dalzellia species. Further detailed studies on morphology using the protologue made it clear that the plant specimen is Dalzellia ubonensis M. Kato (2006:10).
Discussion
The present study marks the first confirmed report of Dalzellia ubonensis M. Kato from Kerala, India, thereby extending the known geographic range of the species beyond its previously documented distribution in Southeast Asia, including Thailand, Vietnam, and Laos.14 This finding is significant in the context of Podostemaceae biogeography, as it underscores the potential for overlooked or undocumented diversity within Indian aquatic ecosystems, particularly in the Western Ghats region, a recognized biodiversity hotspot.
The taxonomic identification of D. ubonensis from the Kallar River was substantiated through a combination of morphological traits and molecular evidence. Morphologically, the collected specimens exhibited a crustose to foliose shoot system, subdimorphic leaves, and floral characteristics that are congruent with the original protologue of D. ubonensis. 14 These features contrast with those of Dalzellia gracilis, the only other species of the genus previously reported from India.6 In particular, the presence of foliose shoots and the distinctive floral morphology—such as the shape and arrangement of tepals and stamens—clearly differentiate the Kerala specimens from D. gracilis, which is characterized by a more gracile habit and differing reproductive structures.
Comparative molecular analysis, using the MATK gene as a barcode marker, further reinforced species-level identification. The high sequence similarity (≥98% identity) and complete query coverage in BLASTn analyses against verified D. ubonensis sequences provide strong phylogenetic support for the identification. This integrative approach aligns with recent trends in aquatic plant taxonomy, where DNA barcoding has become a valuable tool in resolving taxonomic ambiguities, particularly in morphologically plastic or cryptic species.
Ecologically, the presence of D. ubonensis in the Kallar River is consistent with the habitat preferences reported in prior studies from Southeast Asia, where the species is known to inhabit submerged rock surfaces in clear, fast-flowing rivers. 14 The specificity of this microhabitat underscores the narrow ecological amplitude of Podostemaceae members and raises concerns regarding their vulnerability to hydrological alterations. Previous studies have documented that Podostemaceae species are highly sensitive to changes in flow regime, sedimentation, and water pollution, 1 all of which are increasingly common in riverine systems of the Western Ghats due to anthropogenic pressures.
The discovery of D. ubonensis in India has important implications for the regional flora and prompts a re-evaluation of the genus Dalzellia within the Indian subcontinent. It suggests that historical under-sampling or taxonomic oversight may have led to an underestimation of species diversity within this genus. Given that Podostemaceae are often restricted in distribution and exhibit high levels of endemism, our findings call for a more comprehensive survey of aquatic habitats across the Western Ghats and northeastern regions of India, where similar ecological conditions may support undiscovered or unreported species.
Conclusion
In conclusion, this study contributes to the growing body of literature emphasizing the importance of integrative taxonomy in documenting plant diversity. By confirming the presence of Dalzellia ubonensis in Kerala through detailed morphological and molecular evidence, it fills a critical gap in the known range of the species and highlights the Kallar River ecosystem as a significant site for aquatic plant diversity. Future research should focus on population genetics, reproductive ecology, and habitat monitoring to support effective conservation of D. ubonensis and related taxa in India.
Acknowledgement
The authors are grateful to management of NSS College, Pandalam and to the Post Graduate and Research Department of Botany for all the facilities provided.
Funding Sources
The author(s) received no financial support for the research, authorship, and/or publication of this article.
Conflict of Interest
The authors do not have any conflict of interest.
Data Availability Statement
This statement does not apply to this article.
Ethics Statement
This research did not involve human participants, animal subjects, or any material that requires ethical approval.
Informed Consent Statement
This study did not involve human participants, and therefore, informed consent was not required.
Clinical Trial Registration
This research does not involve any clinical trials.
Permission to reproduce material from other sources
Not Applicable
Author Contributions
Fouzia Hilal: Conceptualization, Methodology, Writing – Original Draft, Data Collection, Analysis, Writing – Review & Editing.
Jithesh Krishnan Ramakrishnan Nair: Data collection, Visualization, Supervision, Project Administration.
References
- Philbrick C.T., Novello K.A. New World Podostemaceae: ecological and evolutionary enigmas. Brittonia. 1995;47(2):210-222.
CrossRef - Cook C.D.K. Aquatic and wetland plants of India. Oxford University Press; 1996:385.
CrossRef - Koi S., Ikeda H., Rutishauser R., Kato M. Historical biogeography of river-weeds (Podostemaceae). Aquatic Botany. 2015;127:62-69.
CrossRef - Tang H.T., Kato M. Culture of river-weed Terniopsis chanthaburiensis (Podostemaceae). Aquatic Botany. 2020;166:103-255.
CrossRef - Kato M. Distribution and biogeography of Podostemaceae in Asia. Bull Natl Sei Mus Tokyo Ser B. 2006;32:19-22.
- Cook C.D.K., Rutishauser R. Podostemaceae. In: Kubitzki K, ed. The families and genera of vascular plants. Springer; 2007:304-344.
- Koi S., Rutishauser R., Kato M. Phylogenetic relationship and morphology of Dalzellia gracilis (Podostemaceae, subfamily Tristichoideae) with proposal of a new genus. Int J Plant Sci. 2009;170:237-246.
CrossRef - Fujinami R., Imaichi R. Developmental anatomy of Terniopsis malayana (Podostemaceae, subfamily Tristichoideae), with implications for body plan evolution. J Plant Res. 2009;122:551-558.
CrossRef - Mathew C.J., Jäger-Zürn I., Nileena C.B. Dalzellia gracilis: a new species of Podostemaceae (Tristichoideae) from Kerala, India. Int J Plant Sci. 2001;162:899-909.
CrossRef - Imaichi R., Maeda R., Suzuki K., Kato M. Developmental morphology of foliose shoots and seedlings of Dalzellia zeylanica (Podostemaceae) with special reference to their meristems. Bot J Linn Soc. 2004;144:289-302.
CrossRef - Fujinami R., Imaichi R. Developmental morphology of flattened shoots in Dalzellia ubonensis and Indodalzellia gracilis with implications for the evolution of diversified shoot morphologies in the subfamily Tristichoideae (Podostemaceae). Am J Bot. 2015;102(6):848-859.
CrossRef - Drummond, A.J., Ashton, B., Buxton, S., Cheung, M., Cooper, A., Heled,J ., Kearse, M., Moir, R., Stones-Havas, S., Sturrock, S., Thierer, T. and Wilson, A. (2010). Geneious v5.1, Available from http://www.geneious.com.
- Kato M. Taxonomic studies of Podostemaceae of Thailand. I. Hydrobryum and related genera with crustaceous roots (subfamily Podostemoideae). Acta. Phytotax. Geobot.2004;55(3):133-165.

This work is licensed under a Creative Commons Attribution 4.0 International License.





